AMENDMENT #1 TO CLINICAL STUDY AGREEMENT
Exhibit 10.22
AMENDMENT #1 TO
The following is Amendment #1 to the Clinical Study Agreement dated July 17, 2017 between Volition America, Inc. (“Laboratory”) and the Regents of the University of Michigan (the “University”) for Xxxx Xxxxxxx, ORSP Reference # AWD005981, and incorporates all of the terms therein. This Amendment is effective as of September 1, 2019.
The purpose of this Amendment is to acknowledge that the requirements of the GLNE010 study have been satisfied. This Amendment serves to end the completed sections of the Study Protocol in Exhibit A (GLNE010) add Study Protocol Exhibit A-1 {GLNE007}, Exhibit B and Exhibit C. This Amendment shall also replace Section 3.1 in its entirety:
3.1In consideration of its participation in the Clinical Study on the terms and conditions of this Agreement, Laboratory shall provide direct and indirect funding in the amount of up to Xxx Xxxxxxx, Xxxx Xxxxxxx Xxxxxxxx Xxxxxx Xxxxx Dollars (US$1,500,000). Direct and indirect payments by the Laboratory for the Clinical Study has concluded, are paid in full, and no additional future funding will be provided.
All terms of the Clinical Study Agreement shall remain unchanged.
ACCEPTED AND AGREED TO:
Volition America, Inc.
/s/ Xxxxx Xxxxxxx 02/17/2020
Signature of Responsible Officer for LaboratoryDate
Xxxxx Xxxxxxx, MD, Chief Executive Officer
Typed Name and Title
Regents of the University of Michigan:
/s/ Xxxxx Xxxxxxx 02/11/2020
Signature of Responsible Officer for LaboratoryDate
Xxxxx Xxxxxxx, Project Representative
Typed Name and Title
Evaluation of Stool Based Markers for the Early Detection of Colorectal Cancers and Adenomas
Great Lakes New England (GLNE) Clinical Validation Center
NCI Early Detection Research Network
STUDY INVESTIGATORS:
Clinical Validation Center Investigators (GLNE)
Xxxxx Xxxxxxxx, M.D.1
Xxxx X. Xxxxx, M.D.2
Xxxx X. Xxxxxxx, M.D.1
Xxxxxx Xxxxxxxxx, M.D.3
Xxx Xxxxxx Ph.D.4
Xxxx Xxxxxxxx, M.D., MPH2
Xxxxxx Xxxx-Xxxxxx, M.D., Ph.D.9
Xxxxxxx X. Xxxxx, M.D.8
Xxxx Xxxxxx, M.D., M.P.H5
Ananda Sen, Ph.D.1
Xxxxx Xxxxxxx, M.D., M.P.H.1
Xxxxx Xxxxxx, M.D. 6
Xxxxxxxxxxx Xxxxxxx M.D.7
1University of Michigan Medical Center, Ann Arbor, MI
2University of North Carolina, Chapel Hill, NC
3MD Xxxxxxxx Cancer Center, Houston, TX
4University of Minnesota, Minneapolis, MN
5Hershey Medical Center-PSU, Xxxxxxx, XX0
6Dana-Xxxxxx Harvard Cancer Center, Boston, MA
7Saint Michael’s Hospital, University of Toronto, Toronto, Ontario
8University of Washington Medical Center, Seattle, WA
9University of Puerto Rico Comprehensive Cancer Center, San Xxxx, Puerto Rico
Contact information for Great Lakes New England Clinical Validation Center
2150 Cancer Center and Geriatrics Center
University of Xxxxxxxx Xxxxxxx Xxxxxx Xxx Xxxxx, XX 00000-0000
Telephone: (000) 000-0000
Fax: (000) 000-0000
Email:xxxxxxxx@xxxxx.xxx (PI)
xxxxxxx@xxxxx.xxx (Lead CRA)
EDRN Data Management and Coordinating Center (DMCC)
Xxxx Xxxxxxxxxx Cancer Research Center
0000 Xxxxxxxx Xxx X Xxxxxxx, XX 00000-0000
000-000-0000
Ziding Feng, Ph.D. (PI)
xxxxx@xxxxxxxxx.xxx (DMCC GLNE 007 Project Manager)
GLNE 007
EVALUATION OF STOOL BASED MARKERS FOR THE EARLY DETECTION OF
COLORECTAL CANCERS AND ADENOMAS
1.0 | Summary of Study | 1 |
2.0 | Schema | 2 |
3.0 | Objectives | 3 |
4.0 | Background and Significance | 3 |
4.1 | Biomarkers | 3 |
4.2 | The Early Detection Research Network (EDRN) | 3 |
4.3 | Current State of the Art: Recommended Early Detection | 4 |
4.4 | Current State of the Art: Serum Based Biomarkers for Colorectal Neoplasia | 4 |
4.5 | Rationale and Current State of the Art: Stool Based Biomarkers for Detection of Colorectal Neoplasia | 6 |
4.6 | Key Issues Driving Research Questions in CRC Early Detection Biomarkers | 8 |
5.0 | Study Design | 9 |
5.1 | Summary of Study Plan | 9 |
5.2 | Rationale for tissue collection | 9 |
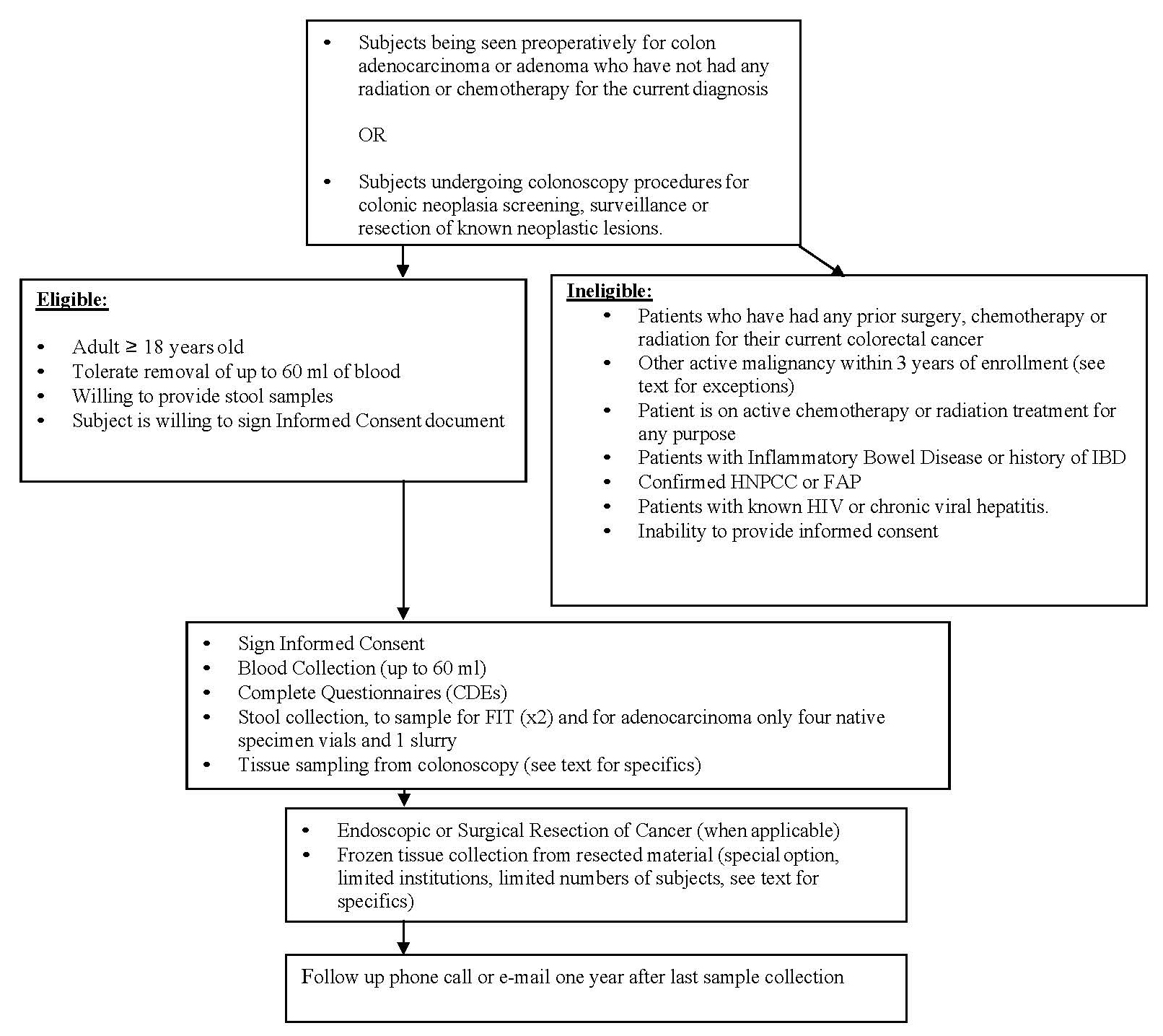
6.0 | Inclusion and Exclusion Criteria | 9 |
6.1 | Inclusion Criteria | 9 |
6.2 | Exclusion Criteria | 11 |
7.0 | Study Procedures | 11 |
7.1 | Subject Recruitment | 11 |
7.2 | Clinical Procedures | 11 |
7.3 | Biological Sample and Data Collection | 13 |
7.4 | Circulating methylated genes BCAT1/IKZF1 (Clinical Genomics) | 14 |
7.5 | Hypomethylated LINE1 from circulating cell free DNA (VolitionRx) | 15 |
7.6 | Disclosure of results to subjects | 15 |
7.7 | Evaluable subjects | 15 |
7.8 | Completion of Study | 15 |
7.9 | Subject Compensation | 15 |
8.0 | Study Calendar | 16 |
9.0 | Statistical Considerations | 16 |
9.1 | Study Population | 16 |
9.2 | Data Analysis Plan | 17 |
9.3 | Justification of Design and Sample Size | 18 |
10.0 | Data Safety and Monitoring | 19 |
10.1 | Data Safety and Monitoring | 19 |
11.0 | Adverse Event Reporting | 20 |
12.0 | Data Management | 20 |
12.1 | Registration | 20 |
12.2 | Timeliness | 20 |
12.3 | Completeness and Accuracy | 20 |
12.4 | Accuracy--Revisions and Corrections | 20 |
12.5 | On Site Data Audits | 21 |
12.6 | Sample Tracking | 21 |
12.7 | Confidentiality | 21 |
12.8 | Security | 21 |
13.0 | Ethical and Regulatory Considerations | 22 |
13.1 | Institutional Review | 22 |
14.0 | References | 23 |
1.0SUMMARY OF STUDY
As part of the National Cancer Institute-funded Early Detection Research Network (EDRN), the Great Lakes-New England Clinical Epidemiological Center (GLNE CEC) proposes a research study that validates potential molecular markers (“biomarkers”) for the detection of precancerous and cancerous conditions and cancer risk assessment. Although examples of such biomarkers are currently in clinical use (i.e. CEA, CA-125), there are limitations to all of them. Our consortium focuses on gastrointestinal neoplasia.
The goals of this phase of the proposed research are:
1.Assessment of the utility of individual stool-based, and serum-based biomarkers for discriminating between patients with adenocarcinomas, patients with adenomas with high grade dysplasia, patients with advanced adenomas defined as adenoma histology of any combination including sessile serrated adenoma, tubulovillous adenoma, villous adenoma, sessile serrated polyp/adenoma, traditional serrated adenoma OR any adenoma ≥1 cm OR three or more adenomas, patients with adenomas that are not advanced, and normal colonoscopy subjects both at normal and high risk for developing colon cancer.
0.Xxxxxxxxxxxx of a panel of markers from those considered in Objective 1 to discriminate, under a number of assumptions concerning prevalence and cost of misclassification, between:
a.(Primary) Subjects with normal colons or non-advanced adenomas versus patients with cancers
b.(Secondary) Subjects with normal colons versus patients with cancers.
3.Comparison of the characteristics of individual markers and panels as discriminators to those of the established current standard, fecal immunochemical test (FIT).
4.Development of a bank of stool samples linked to serum, tissue, and clinical data from patients with colorectal cancer, adenomas and normal controls for validation of stool-based markers that may be developed in the future.
To build our collection, we propose to collect stool, FIT, serum, plasma, and tissue samples from 1,000 new subjects. EDRN Common Data Elements (CDEs) will be completed by the recruiting sites and provided for the laboratories developing the assay. Each biomarker will be analyzed individually and considered as a potential panel marker to be used for future large-scale screening longitudinal trials.
This protocol had previously recruited subjects from January 2006 to June 2010. The samples from this recruitment period are 9-13 years old as of the development of this protocol in April, 2019. Prior recruited subjects:
262 adenomas (54 of those advanced)
191 cancers
65 high risk, colonoscopic normal
164 colonoscopic normal
From each subject, we collected 30 serum, 30 plasma, 5 stool, 20 5- ml urine aliquots
Current Status of GLNE 007 repository:
Total circulating space samples collected: 16,900 serum, 15,700 plasma, 3,000 stool, and 7,000 urine aliquots (42,600 total)
Total circulating space samples disbursed: 12,600 aliquots of various types.
Remaining in the collection: (total 30,000)
10,200 Serum aliquots
11,500 Plasma aliquots
2,100 stool aliquots (representing 585 unique subjects with at least 1 aliquot left)
6,200 urine aliquots
Total tissue samples: 2,100 tissue pieces snap frozen in liquid nitrogen
Total tissue samples disbursed: 1,050
Total tissue samples remaining: 1,050
This amended protocol (version 7) proposes to restart GLNE 007 to recruit 1,000 new subjects, (400 colorectal cancers, 200 adenomas, 200 higher risk but endoscopically normals and 200 endoscopically normal colons for controls). Thus, bringing our total from 682 to 1,682 total subjects.
1
NOTES:
Nursing women who otherwise meet the eligibility criteria may participate.
Subjects who had CRC that was successfully treated at least three years ago are eligible.
Recent screening colonoscopy (within 3 weeks of enrollment), poor preparation found at colonoscopy and returning for repeat colonoscopy are eligible.
2
3.0OBJECTIVES
1.Assessment of the utility of individual stool-based, and serum-based biomarkers for discriminating between patients with adenocarcinomas, patients with adenomas with high grade dysplasia, patients with advanced adenomas defined as adenoma histology of any combination including sessile serrated adenoma, tubulovillous adenoma, villous adenoma, sessile serrated polyp/adenoma, traditional serrated adenoma OR any adenoma ≥1 cm OR three or more adenomas, patients with adenomas that are not advanced, and normal colonoscopy subjects both at normal and high risk for developing colon cancer.
0.Xxxxxxxxxxxx of a panel of markers from those considered in Objective 1 to discriminate, under a number of assumptions concerning prevalence and cost of misclassification, between:
a.(Primary) Subjects with normal colons or non-advanced adenomas versus patients with cancers;
b.(Secondary) Subjects with normal colons versus patients with cancers.
3.Comparison of the characteristics of individual markers and panels as discriminators to those of the established current standard, fecal immunochemical test (FIT).
4.Continued support of a renewal of a bank of stool samples linked to serum, tissue, and clinical data from patients with colorectal cancer, adenomas and normal controls for validation of stool- based markers that may be developed in the future.
4.0BACKGROUND AND SIGNIFICANCE
4.1Biomarkers
Definitions, underlying assumptions, and rationale.
A biomarker is defined as a characteristic that is measured and evaluated as an indicator of normal biologic processes, pathogenic processes, or pharmacologic responses to therapeutic interventions (3). An NCI Working Group further characterized a biomarker as a Clinical Endpoint--a characteristic or variable that measures how a patient feels, functions or survives; as a Surrogate Endpoint--a biomarker intended to substitute for a clinical endpoint in a clinical trial-- and as a Global Assessment: an evaluation of risk and benefit balance for a patient or group of patients. However, the working group did not address biomarkers specific to the carcinogenesis process or for cancer detection.
The underlying assumption of a surrogate endpoint for cancer prevention is that a measured biological event will predict a cancer outcome, either immediately or at a later time (4) and, in the same circumstances, be affected by the intervention. The primary motivations for development of such surrogate endpoints concerns the ability to diagnose cancer at an early stage, to identify individuals at high risk for development of cancer and to enable reduction of sample size and trial duration for an interventional trial such that a rare or distal endpoint can be replaced by a more frequent and more proximate endpoint (5).
Specifications for a useful biomarker.
An “ideal” biomarker will have the following characteristics (6):
(i)Variability of expression between phases of the carcinogenesis process (i.e. normal, pre- malignant, malignant).
(ii)Detectable early in the carcinogenesis process.
(iii)Associated with the risk of developing cancer or the occurrence of pre-cancer.
(iv)Detected in body fluids (e.g. blood, urine, sputum) or tissues obtained via biopsy.
(v)Capability for development of adequate quality control procedures.
(vi)Potential for modification by a chemo preventive agent.
4.2The Early Detection Research Network (EDRN)
The mission of the Early Detection Research Network.
The Early Detection Research Network (EDRN) is a comprehensive effort supported by the NCI to develop highly sensitive, specific, and clinically reliable early detection tools. The Network is harnessing scientific expertise from national and international institutions to identify and validate molecular markers for the detection of precancerous and cancerous cells and to assess risk for developing cancer.
0
Xxxxx Xxxxx Xxx Xxxxxxx Clinical Validation Center (GLNE CVC)
The GLNE CVC is a funded EDRN consortium dedicated to the characterization and validation of biomarkers for the early detection and risk assessment of colorectal adenocarcinoma. The consortium provides the EDRN with expertise in population epidemiology, biostatistics, pharmacology and medical oncology.
4.3Current State of the Art: Recommended Early Detection
Randomized controlled trials have shown that annual or biennial fecal occult blood tests (FOBT) reduce colorectal cancer (CRC) mortality by 15% to 33% (7-9). The reduction is durable over 3 decades (10). Population based cohort studies of colonoscopic screening demonstrate reduced CRC mortality, primarily in distal but not in the proximal colon (11-13). This discrepancy has been attributed to endoscopic quality issues, the technical difficulties in detecting lesions in the right colon, and the more frequent occurrence of flat and depressed dysplastic lesions in the right colon (14-17). In tandem colonoscopy studies, a subset of large polyps may be missed by a single examiner. Shorter withdrawal is time-linked to a lower adenoma detection rate (18, 19). Flat and depressed lesions are more challenging to detect and have been described with a relatively high prevalence in a US colonoscopy cohort (20). While colonoscopic removal of adenomatous polyps reduces CRC mortality (21), prospective, randomized controlled trials of screening colonoscopy have been initiated by the VA and in Europe (21-23). Over-diagnosis (i.e. early detection of indolent invasive neoplasms that do not cause mortality) or lead-time bias in early detection of colorectal neoplasms do not degrade the efficacy of screening and early detection for colorectal cancers (24).
Current screening guidelines for average risk individuals vary world-wide. In the United States the American Gastroenterology Association recommends testing for early detection of adenomas and cancer (structural examination) or of cancer (non-invasive stool tests) beginning at age 50 (25). The United States Preventive Services Task Force (USPSTF) recommends fecal occult blood testing (FOBT) every two years with optional endoscopic screening with either flexible sigmoidoscopy or colonoscopy (26). The majority of developed countries recommend fecal occult blood testing every two years but do not support endoscopic screening (27); albeit with some exceptions (e.g. Germany (27, 28)). In 2012, 65.1% of the United States adults adhered to USPSTF colorectal screening guidelines with colonoscopy the commonly used screening method (61.7%) followed by FOBT (10.4%) (29) whereas colonoscopic screening adherence in Germany is 16% (28). Over 20 years of SEER data (1991 to 2011), United States CRC incidence (all races, males, females) has fallen from 59.5 cases in 1991 to 39.3 cases per 100,000 in 2011 (35% reduction) with a corresponding mortality reduction over the same time period from 24.0 to 15.1 deaths per 100,000 (37% reduction) (30). Widespread adherence to screening guidelines in the United States may be driven by the profound changes in the organization of medical care including enhanced access via the Affordable Care Act, rigid guideline enforcement by payers with physician performance incentives and disincentives, and the rapid adaptation of electronic medical record systems enabling ease referrals for screening, compliance reminders, and management tracking of compliance to care guidelines (31).
4.4Current State of the Art: Serum Based Biomarkers for Colorectal Neoplasia
Rational for non-adherence with stool based or colonoscopic based CRC screening include the volume of bowel preparation, inadequate analgesia, no recommendation from primary physician, embarrassment (32) or cultural taboos surrounding collection or manipulation of stool provide rationale for discovery and validation of circulating biomarkers for early detection of colorectal neoplasia. Circulating signatures may be detected from neoplasm generated genetic products, antigens, antibodies, glycans, circulating tumor cells.
4
Genetic Products
In a recent study of 24 CRC patients, mutant DNA fragments (circulating tumor DNA, ctDNA) are found in relatively high concentrations in the circulation of most patients with metastatic cancer and at were detected in ~70% of patients with localized cancers (33). The direct detection of aberrant genes or genetic material specific to colorectal neoplasms (e.g. APC, b-catenin, K-ras, DCC, and p53) has been limited by the technical challenge of DNA recovery, the large number of potential underlying genetic mutations, and by the limited sensitivity of any single genetic alteration due to the extremely low abundance gene mutations in circulating plasma or serum (33- 38). DNA hypermethylation, in contrast, affects residues in regulatory portions of genes and provides major advantages in designing biomarker assays (37, 39-41). Digital based quantitative technologies improving upon bisulfite conversion while minimizing bisulfite associated DNA fragmentation and single molecule detection technologies (42) permit cost effective development of DNA hypermethylated gene biomarkers. Such technology detected circulating methylated vimentin with 59% sensitivity (42). Septin9, a methylated gene discovered in tissues with array technology (43, 44), detects CRC with 50% sensitivity and 92% specificity in a large (7941 participants) prospective colonoscopy verified screening trial (45). For early stage CRC, Septin9 sensitivity decreased to 35%. While circulating methylated CpG DNA promoter sites appear to have higher CRC detection performance than other genetic detection strategies, they substantially lag behind stool based detection of blood DNA markers or endoscopy. Nevertheless, for individuals refusing to use stool based screening, detection sensitivity of circulating methylated DNA markers appears equivalent to guaiac based stool screening and has the potential advantage of capturing the 40% of the population refusing stool screening. miRNAs are stable and detectable in serum and plasma. As in stool, numerous up and down regulated xxXXX stool signatures discovered using unsupervised array technology may be useful as CRC detection biomarkers. A recent review identifies 19 miRNAs as individual or groups in panels as candidates for detection markers; but, insufficient clinical validation renders the data generated to date using small convenience sets confusing and not mechanism driven (46).
Proteins
Antigens: Approximately 50% of all proteins are estimated to be glycosylated (47). Glycan abundance and their micro- and macro-heterogeneity can be changed in a disease-specific manner (48). Glycoprotein screening studies, many EDRN supported, have relied on immunoprecipitation or lectin affinity capture of whole glycoproteins and mass spectrometry identification of the de- glycosylated protein portion or probed with lectins in an array format containing up to a few hundred antibodies (49-53). Sialylated Xxxxx A and Xxxxx X moieties carrying proteins identify panels of potential markers. The Xxxxx EDRN laboratory has found seven such proteins (B3GNT5, CD44, HSPG2, IL6, INHBC, NOTCH4 and VWF) which when combined in discovery set plasma samples ROC AUC of 0.83 (54). GLNE discovered glycan ligand, galectin- 3 ligand is a circulating glycan biomarker in large population based prospective validation (55).
Antibodies: Serum antibodies recognizing multiple colon cancer antigens can be detected in colorectal adenocarcinoma patients’ markers (56-58). Preliminary validation of single or small autoantibody panels have been disappointing (59). For example, antibodies to the Fas receptor have 17% sensitivity when 100% specific for CRC detection (60). Experience with p53, Hsp60, and nucleobindin 1 (Calnuc) autoantibodies has been better (~50% sensitivity/70 to 90% specific); but, they are not specific to CRC (59, 61, 62) and cannot be used as a colon specific screening tool. Discovery sets that include a miniarray of autoantibodies with other markers have reported improved detection accuracy (sensitivity 83%/specificity 90%) (63) but require clinical validation.
Cytokines/growth factors: High serum concentrations of insulin-like growth factors (IGF) and low levels of their binding proteins have been shown to correlate with CRC risk in large cohort studies (64-67) but have low sensitivities with high specificities for CRC detection. Other cytokines or angiogenesis factors such as TGF-b1 (68-74), VEGF (75, 76), angiogenin (77), endostatin (78), and endothelins (79, 80) also have low sensitivity in small convenience sets and have not proceeded to clinical validation.
Other proteins: Of the matrix metalloproteinases (81-83), plasma TIMP1 is elevated in CRC but has not had sufficient sensitivity in larger validation trials to merit development as a detection biomarker (84). Cell adhesion molecules (85) have low sensitivities for detection of early stage CRC.
5
Circulating Tumor Cells
Circulating tumor cells (CTCs) entering the vascular space from primary neoplasms have been considered to be initiators of metastases (86-88) and can be detected in early stage invasive neoplasms (89, 90). CTC isolation from epithelial cancers initially used antibody capture technology dependent upon epithelial adhesion (EpCAM) and cytokeratins (86). This technology limits CTC detection of early stage neoplasms because CTCs are thought to undergo epithelial to mesenchymal transition (EMT), epithelial traits are lost and epithelial marker such as EpCAM and cytokines are downregulated. CTCs present in as few as 1 cell in 5 x 109 red cells, and up to 5–10 x 106 white blood cells, are rare events (88). Newer microfluidic or centrifugation devices appear to more efficiently capture CTCs (89, 91). The inclusion of mesenchymal/EMT-specific antibodies, for example, vimentin, PLS3 may improve CTC capture and/or expansion (88). With the emergence of ex-vivo expansion protocols of CTCs and the increased ability to detect stem like or stem progenitor cells, CTCs are of future interest as an early cancer detection diagnostic (89, 91), but remain in the technology development phase.
Special consideration—EDRN discovered and preliminarily validated circulating biomarker: Galectin-3 Ligand ELISA as a Serum Biomarker for the Detection of Colorectal Neoplasia
The galectins are widely distributed and evolutionarily conserved carbohydrate binding proteins characterized by their binding affinity for β-galactosides and by conserved sequence elements in the carbohydrate-binding region (92). Galectin-3 is the galectin that is of most interest in regard to colon cancer because of its demonstrated role in cancer progression, metastases, and interaction with mucins(93-97). Galectin-3 ligands include laminin, LAMP-1 and 2, LPS and colon cancer mucin. The major galectin-3 ligand detected in serum is a 40 kDa band distinct from MUC2 and other mucins CEA, and Mac-2-BP. We reported a true positive rate for the detection of CRC of 91% and false positive rate of 18% using preliminary data using quantitative Western blot technology on a convenience set of GLNE serum (55).
We developed a sensitive, reproducible ELISA assay for galectin-3 using a new antibody we created. This was used to assay the GLNE colorectal reference set (50 colorectal adenocarcinomas/50 adenomas/50 endoscopically normal controls). The ROC analyses for galectin-3 ligand combined with FOBT (fecal occult blood test-guaiac based) for detection of colorectal adenocarcinoma versus controls who had normal colonoscopy shows an area under the ROC curve of 0.91, while galectin-3 ligand detection of colorectal adenocarcinoma alone versus controls who had normal colonoscopy shows an area under the curve of 0.84. The true positive rate of galectin-3 ligand with FOBT for detection of CRC is 64% with a false positive rate of 5%. Without FOBT, true positive rate of galectin-2 ligand was 72% with a false positive rate of 20%.
4.5Rationale and Current State of the Art: Stool Based Biomarkers for Detection of Colorectal Neoplasia
Occult blood tests
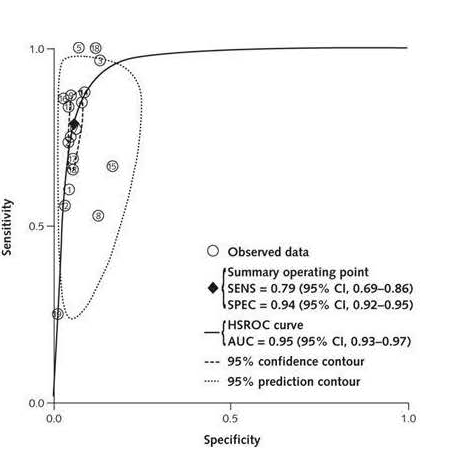
Stool testing as a screening approach offers the potential advantages of noninvasiveness, low cost, avoidance of cathartic preparation, and minimal impact on work time or daily activities. Guaiac based FOBT is not specific for human blood, and consequently, it has a high false positive rate for colorectal neoplasia. The fecal immunochemical test (FIT) detects human hemoglobin, thus eliminating the false positives caused by non-human hemoglobin in the diet (98, 99). FIT tests are more sensitive at detecting CRCs (sensitivity range 61% to 91%) and adenomas (sensitivity range 16% to 31%) than classical unrehydrated guaiac FOBT (Hemoccult II) (sensitivity range 25% to 38% for CRC; 16% to 31% for advanced adenomas) (100, 101). A recent meta- analysis that analyzed data from 19 prospective randomized trials or cohorts using 8 different commercially available FIT tests with colonoscopy or 2-year observation endpoints reported an overall sensitivity for detection of CRC of 79% (95% CI = 0.69-0.86), specificity of 94% (95% CI = 0.92-0.95) and overall accuracy (defined as hierarchical summary receiver operating characteristic (ROC) curve) of 95% (95% CI = 93% - 97%) (Figure 1). Differences in performance characteristics among FIT brands were small, particularly between the two major brands used OC-Light (Eiken Chemical) and OC-Micro/Sensor (Polymedco + Eiken Chemical). The Polymedco product is widely used in the USA. Quantitative FIT (Eiken OC-SENSOR) >177 µg/gm stool combined with age and sex predicts 11.46 fold risk of a large adenoma over lower risk groups (102).
6
| Fig 1 from Xxx et al (1): Hierarchical ROC curve of the sensitivity versus specificity of FIT. The diamond = summary point of the curve to which the pooled sensitivity and specificity correspond. Dashed line = 95% CI for summary point; dotted line = 95% confidence area of FIT diagnostic accuracy. AUC = area under the curve; SENS = sensitivity; |
Stool DNA tests
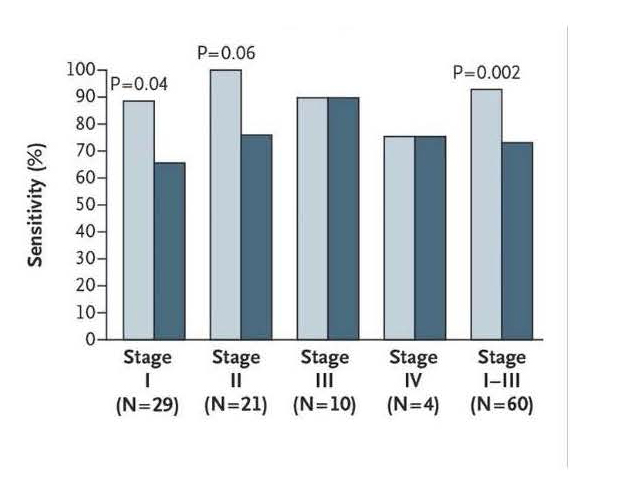
Since the neoplastic transformation process of the colonic epithelium results in cells shedding into the stool, collection of fecal material is likely to yield detectable molecular and biochemical events associated with cellular transformation (103, 104). First generation multi-marker stool DNA tests detected 52-73% of CRCs, 41-49% of CRCs plus adenomas with high grade dysplasia, and 15-46% of adenomas ≥1 cm, with specificities of 84- 95% (105, 106). Stool DNA test performance in both studies was compromised by failure to use stabilization buffer with stool collection, inefficient marker recovery from stool, and relatively insensitive analytical methods. Exact Sciences modified their previously published stool DNA panel (106) and now uses a panel consisting of methylated BMP3 and NDRG4 promoter regions, mutant K- ras (7 point mutations, Exon 2, codons 12,13), and a proprietary FIT test. In a recently published cross sectional validation study of 9,989 patients undergoing screening colonoscopy, the panel performed with a sensitivity of 92% for CRC; 84% for CRC + high grade dysplasia; and 42% for advanced adenomas (Figure 2) (2). The specificity was 87% for CRC, the ROC AUC for the Exact Sciences DNA stool panel for the detection of colorectal cancer is 0.94. FIT alone (Polymedco FIT) performed with sensitivity of 73.8% and specificity of 94.9% for detection of CRC and sensitivity of 23.8% for screen relevant neoplasia. Stool DNA component of the panels adds ~20% sensitivity to FIT. The USPSTF is currently assessing the role and contribution of fecal DNA panels such as the Exact Sciences panel to CRC screening (107).
| Fig 2 from Imperiale et al (2) sensitivity for detection of CRC by Exact Sciences stool DNA panel + FIT (light blue) vs Fit alone (dark blue) by stage. |
7
Vimentin Methylation as a Stool DNA Test
Aberrant methylation of vimentin exon 1 was initially described as a highly frequent biomarker of colorectal cancers and adenomas by Xxxxxxxxx and co-workers (108). In reproducible studies, aberrant methylation of vimentin has been detected in 72%-83% of colon cancers and 70%-84% of colon adenomas (108, 109). The current assay for detection of vimentin exon 1 methylation is based on using methylation specific PCR (MSP). Adaptation of the vimentin MSP to testing fecal DNA is accomplished by recovery of vimentin DNA sequences from human stool using hybrid capture to vimentin specific oligonuclotides (108). Initial study showed that MSP assay of vimentin purified from feces (fecal vimentin DNA) detected methylated fecal vimentin DNA in 46% of cancer patients (N=94) at a specificity of 90% (N=198)(108). This initial study involved collaboration between the Xxxxxxxxx laboratory who had discovered the methylated vimentin DNA marker, and Exact Sciences, who implemented detection of this marker in fecal DNA. This initial study was limited by use of samples that had suffered problems of DNA degradation during sample collection and shipping (106). A recently published two stage follow-up study lead by Xxxxxxxxx et al in collaboration with Exact Sciences and the Xxxxxxxxx laboratory showed markedly improved results with the use of a DNA stabilizing buffer added to stools at the time of collection (110). Detection of methylated fecal vimentin DNA was found in 77% of cancers (N=82) at 83% specificity (N=363). Six of 7 adenomas with high-grade dysplasia were also detected. This assay has successfully detected 55% (N=22) of adenomas that were greater or equal to 1cm in size (110). This is a published assay of capture of fecal vimentin DNA and then MSP detection of methylated vimentin exon 1 sequences (108, 110, 111).
Other Stool Based Biomarkers Under Investigation
Considerable interest in fecal microbiome populations has triggered EDRN supported investigators into identifying unique bacterial species that are associated with colonic carcinogenesis and suggests that a microbiome signature may be a useful stool biomarker for CRC risk (112, 113). Metabolome signatures promise to identify amino acid or fatty acid profiles associated with colorectal cancer or high risk (114) have been preliminarily developed in EDRN supported research. Micro-RNAs (xxXXX) have both oncogenic and suppressor properties, can be detected in stool, and have been explored as stool based early detection biomarkers (115, 116). Studies published to date have used small convenience samples and array technologies that have identified diverse and non-reproducible miRNAs as classifiers for colonic neoplasms.
4.6Key Issues Driving Research Questions in CRC Early Detection Biomarkers
Until therapeutic agents with much greater potency and minimal side effects are developed, the current best strategy for reducing cancer morbidity and mortality is early detection of neoplastic disease (117). Key opportunities in the current state of colorectal screening and early detection include:
1.Enhancing adherence to current screening guidelines: Screening and early detection reduce mortality from colorectal cancer; yet 35% of the population in the USA remain non-adherent.
Adherence is much lower in other countries (28). The barriers to these recommendations (cost, discomfort, cultural taboos) may be overcome with circulating biomarkers that provide individuals with persuasive evidence that undergoing invasive screening procedures, i.e. colonoscopy, will have important life-saving benefit that reduces mortality from CRC (11-13, 21). Developing, validating and bringing circulating biomarkers to population screening use remains a high priority that will likely increase adherence to endoscopic screening.
2.Tailoring colonoscopic screening to individual risk: Recently published data from the Clinical Outcomes Research Initiative found the prevalence of large polyps higher in blacks than whites among both men and women (118). Tailoring endoscopic screening to those at risk while limiting screening for those with minimal or no risk (119, 120) will enhance screening adherence and eliminate excess cost. Recommendations for tailoring were primarily population demographic based (119, 120); yet, the translation of carcinogenesis biology and genetics into biomarker panels with extremely high sensitivity (99%), i.e. no false negative tests, promises precise tailored endoscopic screening. The current state of art stool using based biomarker tools is coming close—92% sensitivity (2) but insufficient to permit tailored or individualized risk.
3.Persistently positive stool DNA tests with negative colonoscopic screening: The stool methylated DNA panel’s report 5% false positives (2, 111). A positive stool DNA test with a negative screening colonoscopy could potentially arise from neoplasia in the upper gastrointestinal tract or from occult and missed lesions in the colorectum. The latter is a particular concern in the right colon, where flat lesions and/or sessile serrated adenomas are more prevalent. Preliminary data from the Case Western EDRN BDL found near 100% vimentin methylation in gastric dysplasia while no methylation in adjacent gastric mucosa (X. Xxxxxxxxx, Personal Communication). In Xxxxxxx’x esophagus (BE), 7 of 7 high grade dysplasias (HGD), and 15 of 18 esophageal adenocarcinomas (EAC) and even in some squamous cancers (SCC) had methylated vimentin, whereas it was absent in all 9 normal squamous mucosa (121). A “false positive” stool DNA test may detect dysplasia or invasive neoplasms in the upper GI tract.
8
5.1Summary of Study Plan
We propose a multi-center, prospective, cross-sectional cohort validation study of 1,682 subjects. We propose to increment the original GLNE 007 cohort with 400 subjects with diagnosed colorectal cancer, 200 subjects with colorectal adenomas, 200 subjects a prior history of adenomas, colorectal adenocarcinoma (>3 years previous), returning for surveillance or positive stool test (DNA or blood) but have a normal colonoscopy (higher risk normal), and 200 subjects who have a normal colon with NO prior history of adenomas, colorectal adenocarcinoma (not returning for surveillance) and who do not have a current (within 12 months) positive stool test (DNA or blood) (normal risk) and have a normal colonoscopy . Subjects will be recruited as described in Appendix B. The baseline visit should be done prior to a scheduled colonoscopy. If a subject is suspected of having a colon adenocarcinoma or an adenoma, the baseline samples should be collected before any procedure to remove the cancer or adenoma so the lesions are present when sample collection is done. Patients with cancer must have their baseline visit and all sample collection completed prior to endoscopic or surgical resection of CRC and chemotherapy and/or radiation therapy. Informed consent, demographic information and medical history via questionnaires, blood, and collection will be done at baseline. Stool collection, to sample for FIT (x2) and for adenocarcinoma four native stool specimen vials and 1 slurry will be done as described in the study calendar and Appendix D. All samples will be collected, handled, transported, processed, and stored according to detailed standard operating procedures and will be de-identified by random Specimen ID linked to the Participant ID in VSIMS. Selected subjects, based upon estimated future biomarker requirements, will have normal colonic epithelium collected during the colonoscopy procedure for future biomarker research. For those subjects with a large adenoma found on endoscopy, a frozen biopsy will be requested. Cancers, for the most part will be identified following endoscopic diagnosis.
5.2Rationale for tissue collection
A primary goal of GLNE 007 is to provide biosamples for training and testing of biomarkers that the EDRN believes have potential for future validation for regulatory review. A secondary goal of GLNE 007 meets the EDRN’s discovery and early phase characterization of biomarkers. The EDRN is a vertically integrated organization that includes laboratories doing discovery research and early detection performance characterization research. The GLNE supports all of the EDRN’s missions—discovery, characterization, training and testing in addition to large scale regulatory validation.
On occasion, investigators need fresh tissue to develop and test new biomarker technologies. The GLNE maintains a repository of frozen normal and adenomatous biopsy samples for this purpose. As with other biosamples proposed for GLNE 007, the frozen tissue samples need revitalization and updating.
GLNE collects fresh biopsies from adenomas as made available by local pathologists. If available, GLNE will also collect fresh biopsies from invasive cancers at endoscopy.
Biopsy tissue samples are not required from every subject entered into GLNE 007. The GLNE collects biopsies from normal colonic mucosa from subjects undergoing colonoscopy who are found to have a normal exam on an as needed basis (approximately 10% or 100 subjects will be asked to undergo biopsy of normal colonic mucosa) with a small repository, to be made available to EDRN investigators for discovery and early phase characterization. Because of the extra risk, time for participants and extra costs to the GLNE involved with performing these biopsies endoscopically, we do not require all patients with normal colonoscopies to undergo biopsy and tissue collection of normal colonic mucosa. The GLNE pays centers extra beyond the usual costs to procure frozen biopsy samples. Normal biopsies may be used as controls for EDRN laboratory biomarker discovery research controls for comparison with adenoma tissue and invasive neoplasm tissue. GLNE 007 has provided this resource to the EDRN over the last 15 years and continues to do so.
6.0INCLUSION AND EXCLUSION CRITERIA
6.1Inclusion Criteria
Willing to sign informed consent
Able to physically tolerate removal of up to 60 ml of blood
Adults at least 18 years old
Willing to collect 2 stool samples to prepare FIT test (x2) and for adenocarcinoma 4 native specimen vials and 1 slurry
Nursing women who otherwise meet the eligibility criteria may participate
Subjects undergoing colonoscopy for screening or surveillance (known prior neoplasms resected).
9
Screening Colonoscopy
No known colonic neoplastic disease. Undergoing colonoscopic screening based upon current colon cancer screening guidelines.
Subjects whose screening colonoscopy shows any of these types of polyps may be included in the normal or the higher risk normal bin if they meet the other criteria noted above.
Hyperplastic polyps
Benign mucosal polyps
Polypoid granulation tissue
Prolapsed mucosal polyps
Inflammatory polyp
Transitional mucosal polyp
Lipoma
Gangleoneuroma
Neuroma
Hamartomatous polyp
Subjects who had colorectal adenocarcinoma that was successfully treated at least three years prior are eligible.
Recent screening colonoscopy (within 3 weeks of enrollment), poor preparation found at colonoscopy and returning for repeat colonoscopy.
Recent diagnostic colonoscopy (within 3 weeks of enrollment) with detection of adenocarcinoma or adenoma.
Known colorectal adenocarcinoma or adenoma remains in place after a diagnostic colonoscopy— adenocarcinoma or adenoma in colon at time of blood and stool collection.
Enrolled participants will be grouped into Bins according to one of the following:
Colorectal Cancer-pathologically confirmed colorectal cancer either present at time of stool collection or discovered during colonoscopy (Cancer Bin)
Adenoma-pathologically confirmed adenoma (Adenoma Bin)
Higher Risk Normal (normal colonoscopy)
Negative study colonoscopy and:
oSubjects with a personal history of adenomas (confirmed by pathology) with none present on qualifying colonoscopy
oSubjects with a personal history of CRC (longer than 3 years ago because of exclusion criteria of cancer within last 3 years) with none present at time of qualifying colonoscopy
oAny family history of CRC (1st degree relative)
oCurrent positive screening stool test for blood, for DNA or for both within 12 months.
Normal Control (normal colonoscopy)
Negative colonoscopy and:
oNo prior history of adenomas
oNo prior history of CRC
oNo family history of CRC
oNegative screening test (if performed) for blood, for DNA or for both within 12 months.
10
6.2Exclusion Criteria
Cancer patients who have had any surgery, radiation, or chemotherapy for their current colorectal cancer prior to collecting the baseline samples
Other active malignancy within 3 years of enrollment except any of the following:
a.Squamous cell carcinoma of the skin
b.Basal cell carcinoma of the skin
c.Carcinoma in situ of the cervix, Stages Ia or Ib invasive squamous cell carcinoma of the cervix treated by surgery only. (Excluded if had pelvic radiation)
d.Stage Ia Grade 1 adenocarcinoma of the endometrium treated with surgery
Patient is on active chemotherapy or radiation treatment
Patients with a history of or clinically active Inflammatory Bowel Disease
Patients with known HNPCC or FAP
Subjects with known HIV or chronic viral hepatitis
Inability to provide informed consent
Women who are pregnant
7.1Subject Recruitment
Patients diagnosed with colorectal cancer and adenomas and scheduled for surgical or endoscopic resection or subjects scheduled for a colonoscopy will be recruited from collaborating consortium centers.
The clinical research associate or study nurse (CRA) at each clinical site will identify subjects with appointments for colonoscopy, surgery, endoscopic polyp or cancer removal, or oncology. The study team will obtain permission to review the schedules from the physicians and the Institutional Review Boards. If the physician agrees that their patient can be contacted regarding participation, the research coordinator will meet with the patient in person or send a letter to the patient describing the study. Advertisements (e.g., newspapers, clinics) may also be used to recruit subjects from the surrounding communities.
The letter to the subject will include an opt-in response card. If we receive permission from the subject to contact them, the CRA will discuss the overall study with the potential subject, and arrange for a baseline visit to get consent, baseline samples, and provide stool kit for FIT and specimen vials.
Enrollment and Registration Procedure
Eligible subjects will be enrolled into the study after providing informed consent to analyze stool samples and FIT, and blood samples for biomarkers, medical records review, and for completion of questionnaires. The subject will be assigned a Participant ID by the recruiting site and documented in VSIMS.
Timing of Sample Collection
7.2.1.1Sample collected prior to colonoscopy procedure
Baseline samples, including stool, blood, and FIT must be collected prior to any colonoscopic preparation procedure.
11
7.2.1.2Samples collected with known diagnosis of unresected, untreated colon adenocarcinomas or adenomas.
If any subjects are eligible to begin the study after their colonoscopy (e.g., a lesion remains in the colon), at least 7 days must elapse from the diagnostic colonoscopy, but no more than 3 weeks. Eligibility for the respective bins will be determined from the pathology and colonoscopy reports. Cancer patients must have a diagnosis of colon or rectal adenocarcinoma that has been previously untreated. Any stage is allowed. The baseline stool, FIT, and blood samples must be collected before any surgical resection or chemotherapy or radiation therapy is performed.
Baseline Visit
Informed consent will be obtained prior to any data or sample collection. Samples will be collected either prior to colonoscopic preparative procedure or 7 days or more after a diagnostic colonoscopy as outlined in Section 7.2.2.2. Detailed instructions will be provided to the subjects on the collection of the stool for the FIT tests and for adenocarcinoma four native stool specimen vials and 1 slurry. Samples will be collected as described below. Subjects will prepare two FIT tests and for adenocarcinoma four native specimen vials and 1 slurry from the stool sample for shipping to the University of Michigan.
Data Collection
The subject will be asked to complete EDRN demographic and medical history questionnaires (Appendix A) at baseline. Clarification or additional information may be obtained from the medical records. These data forms have gone through multiple stages of development and testing and are standardized across EDRN studies. Case report forms (CRFs) will also be used to collect information on concomitant medications, colonoscopy outcomes, resection information, cancer treatment, and diagnostics. The Follow up forms and medical record review will be completed at the follow up visit for the subjects in the adenoma and CRC bins if seen in clinic, otherwise done over the phone or e-mail. Long term data collection (medical records review and follow up CDE for all bins) will be done by a phone call or email once at one year post their last contact.
Sample Collection: Blood
Blood samples, up to 60 mLs, will be obtained according to standard operating procedures (Appendix C).
Sample Collection: Stool for FIT Testing
Adenocarcinoma subjects will be provided with a standard collection kit including detailed instructions on how to complete the FIT sampling. All other subjects will only obtain two FIT tests (Appendix D). The first FIT tube will be shipped inside the same shipping container with the stool sample. The second FIT tube will be mailed (pre-paid) to the University of Michigan at room temperature in the manufacturer’s United States Department of Transportation-compliant envelope. The test will be analyzed at the Central Laboratory at the University of Michigan using analytic equipment provided by Eiken Chemical Company. (OC-SENSOR Xxxxx).
Sample Collection: Stool for Biomarker Testing (ADENOCARCINOMA SUBJECTS ONLY)
Subjects with a known diagnosis of colorectal adenocarcinoma will be asked to collect their stool in the collection bucket (hat) provided. Subjects will be given detailed instructions and complete kits to collect the stool samples at home. They will prepare a FIT tube (FIT #1) from the stool sample. Subjects will also collect scoops of stool into a container with an EDTA-based buffer (“buffered stool”) and additional scoops of stool into tubes provided to be sent on ice packs (“native stool”).
The subjects will then package both the stool and the FIT for shipping per provided instructions. The US and Canadian subjects will ship the stool sample to the Central Laboratory at the University of Michigan using pre-paid DOT (Department of Transportation)-compliant packaging. Buffered stool samples will be homogenized and frozen in four 5 ml aliquots at –70° C or colder for batch shipment to the analytical labs. The native stool will be placed at –70° C or colder upon receipt.
Sample Labeling
All samples will be labeled or have an embedded barcode with a unique bar code and linked to the participant ID through VSIMS.
12
7.3Biological Sample and Data Collection
Blood Collection and Storage
Subjects will provide up to 60 ml of blood in six 10 ml collection vials (2 red, 3 purple tops and 1 ACD-A, for serum and plasma respectively). Purple tops tubes must be filled to manufacturer’s level to maintain blood: EDTA ratio. Additional blood draws, prior to prepping for the colonoscopy may be done to get to the necessary blood volume.
The serum samples (red top tubes) will sit at room temperature for a minimum of 30 minutes (maximum of 60 minutes) to allow the clot to form, and if not processed immediately, they can be held at 4° C for a maximum of 4 hours after collection. Plasma samples (purple {aka lavender}top tubes) and ACD-A (yellow top) will be held at 4° C for a maximum of 4 hours after collection. The red top collection tubes will be centrifuged at >1,300 x g at 4° C for 20 minutes (centrifuge brake off for first 10 minutes, then on for last 10 minutes). The serum will be removed, transferred to pre-labeled tubes, and frozen at –70° C or colder. The purple top and ACD-A collection tubes will be centrifuged at >1,300 x g at 4° C for 10 minutes without the centrifuge brake off for first 10 minutes and on for the last 10 minutes. The plasma will be transferred to a 15 ml conical tube for a second centrifugation step (>1,300 x g at 4° C for 10 minutes) prior to aliquoting in pre-labeled tubes, and frozen at –70° C or colder. All frozen samples will be stored at –70° C or colder at the collection site and shipped on dry ice monthly to the Central Laboratory at the University of Michigan and stored at–70° C or colder until assayed. Detailed Standard Operating Procedures including shipping and sample handling instructions are provided in Appendix C.
Stool Sample Collection and Handling (ADENOCARCINOMA ONLY)
Subjects with a known adenocarcinoma will be asked to collect a stool sample at baseline prior to any therapy or resection (when applicable). Subjects will be given a standard stool collection basin (hat) with detailed instructions, shipping container, pre-paid shipping labels, four native specimen collection vials, 1 stool slurry and cold packs, FIT vials, and all necessary supplies.
Subjects will be asked to collect a whole stool sample in the container provided, ensuring that no other materials (e.g. paper or urine) are collected in the hat. Subjects will collect scoops of stool into a container with an EDTA-based buffer (“buffered stool”), additional scoops of stool into tubes provided to be sent on ice packs (“native stool”), and a sample in a FIT vial. The subjects will then package both the stool and the FIT for shipping per provided instructions. Subjects will be asked to prepare four native specimen vials, 1 slurry and FIT tests (x2) (see appendix D) using the materials and instructions provided. The specimen vials will be shipped on the cold packs and frozen at - 80° C at the Xxxxxxx laboratory at the University of Michigan.
Fecal immunochemical Test (FIT) (All enrolled subjects)
Subjects will be asked to prepare two FIT tests (see appendix D) using the materials and instructions provided. The OC-Sensor®, Eiken Chemical Company product, will be used according to manufacturer’s instructions. The threshold for a positive test is 100 ng/ml. The Central Laboratory will process the samples using equipment provided by Eiken Inc. Technicians will undergo tutorial and quality assessment with Eiken support technicians prior to study launch. A quantitative result will be generated and recorded in the database.
Sample Collection: Tissue Samples
NOTE: Tissue sample collection not required for protocol completion. A limited number of tissues per bin will be collected. Collection will be performed at specified designated institutions for incremental payment per accrual
7.3.1.1Collection of Frozen Normal and Adenoma or Cancer Tissue
For individuals with large adenomas who are undergoing endoscopic resection, the fresh surgical sample will be obtained by the endoscopist. Once the adenomas are located, a digital endoscopic picture will be obtained. Once the adenoma (s) is (are) removed, two biopsies will be done or two cuts will be made. The biopsies will then be frozen in liquid nitrogen after being placed in a pre-designated, labeled container. Normal sigmoid tissue will be collected as described below.
Bar coded vials will be sent to University of Michigan sample storage facility. The adenoma will then be sent to the institution’s clinical pathology department according to standard clinical procedures.
13
For cancer or adenoma patients who are undergoing surgical resection, the Site CRA will notify the Pathology Service or the Institutional Tissue Procurement Service of a surgical sample needed for study purposes. Once the specimens are removed, two to four biopsies will be done or two- four cuts will be made. At least one of the biopsies should be from normal colon. The biopsies will be frozen in liquid nitrogen after being placed in a pre-designated, labeled container. Bar coded vials will be sent to University of Michigan sample storage facility. The specimen will be sent to the institution’s clinical pathology department according to standard clinical procedures.
7.3.1.2Collection of Fixed and Frozen Normal Sigmoid Colon Biopsies on Qualifying Colonoscopy
For all subjects who agree to the biopsy portion of the study and are undergoing colonoscopy, the endoscopist will take up to 6 biopsies from the normal sigmoid colon. Of those, at least 2 (and up to 4) will be snap frozen and at least 1 (and up to 2) will be fixed in 10% formalin and sent to the University of Michigan for paraffin embedding by the Histology Core. The fixed and frozen samples will be stored at the University of Michigan GLNE Core Laboratory.
Sample management procedures including storage, tracking, and shipping instructions are provided in Appendix E.
Tissue samples from pathology specimens may be requested for future biomarker studies from samples collected during routine clinical management of patients with adenomas and CRC. Medical records may be re-reviewed to extract data including, but not limited to size and location of tumor, histopathological features, patient treatment, and response to therapy. Patient permission will be obtained via the informed consent document. The University of Michigan core laboratory may request tissue blocks either to cut slides or to keep for future biomarker studies.
Medical Records Documentation
Medical records will be reviewed to collect information regarding the results of the procedures, pathology analysis, surgery, treatment, history, or outcomes and documented in the CRFs/CDEs. The medical records will serve as the source documents and will be maintained at the site enrolling the subject. Since these records necessarily contain subject identifiers, they will not be sent to the Data Coordinating Center at Dartmouth or to the University of Michigan. Medical records may be reviewed at the site during audits or monitoring visits.
Sample labels
All samples will be labeled with bar-coded labels or have embedded bar-codes. The labels will be provided by the DMCC and will link to the subject identification number in VSIMS. Labels will be placed on all tubes in the blood drawing kit, FIT, and frozen stool sample vials.
Sample tracking
All samples will be tracked by a bar code through a computerized program called VSIMS. Upon receipt of the specimen in the University of Michigan Laboratory Core, the bar code will be read and the date and time of arrival, documented. The Data Management Center will be notified at completion of each individual assay performed on a sample.
Long-term Follow up
The CRA will contact the subject via phone or email one year after their last sample collected for additional follow up data. Data will be collected on medical record review and follow up CDE (Appendix A), and include information related to their GI tract history or cancer history and related treatments, procedures, and outcomes. The consent form describes the long-term data collection.
7.4Circulating methylated genes BCAT1/IKZF1 (Clinical Genomics)
A Good Laboratory Practice validated bisulfite PCR assay developed by Clinical Genomics will be used for this assay. Clinical Genomics will perform this assay on blinded samples at their laboratory facility in Rutherford, NJ. Clinical Genomics is not responsible for analysis of any other biomarkers other than their BCAT1/IKZF1 product. Sample distribution schedule is outlined in the Clinical Study Agreement.
14
7.5Hypomethylated LINE1 from circulating cell free DNA (VolitionRx)
A Good Laboratory Practice validated assay developed by VolitionRx will be used for this assay. VolitionRx will perform this assay on blinded samples at their laboratory facility in Namur, Belgium. Volition is not responsible for analysis of any other biomarkers other than their hypomethylated LINE1 assay. Sample distribution schedule is outlined in the Clinical Study Agreement.
7.6Disclosure of results to subjects
Subjects will be informed as part of the consent process that neither they nor their health care providers will receive any results from participation in this study.
A subject is considered evaluable and on-study if all samples are collected per protocol. Subjects without a full set of samples or data may need to be replaced on the study to get 400 evaluable cancers and 200 evaluable subjects the other three bins.
A subject will be asked to provide a replacement sample if:
a.The stool specimens are received outside the time window required (i.e. greater than 36 hours after collection time and/or not kept cold) (Adenocarcinoma subjects only)
x.Xx FIT test
c.Blood cannot be obtained (must be obtained while target lesion is still present for adenomas and cancers)
d.Blood is subject to some kind of handling error (no EDTA, too long at room temperature, etc.) and subject is still eligible to provide the blood again
Protocol deviations
Subjects who do not provide one of the samples or all of the data, but are otherwise eligible to remain on study, will not be reported as deviations.
7.8Completion of Study
A subject has completed the study when the CRF data, blood samples, stool samples (adenocarcinoma only) and FIT have been obtained, properly processed and delivered to University of Michigan, and the one year follow up phone call has been done. A subject may be asked to provide a replacement sample if there is a problem with one collected, including an additional stool sample. The subject may decline, if they choose.
7.9Subject Compensation
To compensate for the inconvenience and cost of driving and parking, $25 will be provided to each subject once blood samples, and stool samples are completed or $50 for adenocarcinoma subjects who have provided all required samples. Recruiting sites will receive gift cards to distribute to subjects that complete the requirements to receive payment. Sites are required to account for distribution of gift cards to subjects. Sites outside the US will receive reimbursement by invoice, instead of gift cards.
15
Procedures |
Baseline1 | Baseline stool collection2 | Colonoscopy/ Resection | Long-Term Follow up |
Eligibility Checklist | X |
|
|
|
Consent Documentation | X |
|
|
|
General Information | X |
|
|
|
Medical History | X |
|
|
|
Concomitant Medications | X |
|
|
|
Colonoscopy |
|
| X |
|
Surgery |
|
| X |
|
Colon Cancer Treatment |
|
|
| X |
Blood Collection | X |
|
|
|
FIT TESTS |
| X2 |
|
|
Stool Sample |
| A2,4 |
|
|
Frozen/Fixed Tissue collection |
|
| S3 |
|
Follow up CDEs |
|
|
| X |
1Baseline clinic visit—Prior to treatment of any colon lesion, or prior to a colonoscopy (specifically, prep), OR at least 7 days post colonoscopy but no later than 3 weeks post colonoscopy.
2Stool collection any time after baseline visit and subject returns home with kits
3Frozen tissue is collected at the time of surgical or endoscopic resection of cancer or colonoscopy findings (fixed and frozen).
4Stool samples in EDTA buffer and 4 vials collected in adenocarcinoma subjects per stool collection and handling SOPs.
S = Special circumstance. Not a required component of protocol completion unless institution is registered as Special circumstance. Additional remuneration provided for a specific number of patients for frozen tissue collection in each of specified bins.
A = Stool collection required for subjects who have adenocarcinoma of the colon or rectum. The patient incentive is increased from $25 to $50 because of their diagnosis and additional effort compared to subjects who do not have cancer.
9.0STATISTICAL CONSIDERATIONS
9.1Study Population
This study is stratified: normal subjects (Stratum 1); subjects at high risk or previously with adenomas who currently are without adenomas (Stratum 2); subjects with adenomas (screen relevant neoplasia (SRN) and non-screen relevant neoplasia (Stratum 3) subjects with colorectal adenocarcinoma (Stratum 4). 200 subjects are to be accrued to each stratum except 400 subjects in Strata 4. Subjects in both Strata 2 and 3 are expected to be more likely to be positive for upstream markers of carcinogenesis than the normal subjects in Stratum 1 (who are both not at high risk and have never had adenomas), while subjects in Strata 3 and 4 are expected to be more likely to be positive for downstream markers indicating the presence of adenomas or adenocarcinomas than those in Strata 1 and 2. Stratum 2 may, therefore, be pooled with Stratum 1 or Stratum 3, depending on the context. From the screening perspective, Stratum 3 will be further divided into SRN or non-SRN. Non-SRN in stratum 3 could be combined with Strata 1&2 to form a non-SRN group and compared to Stratum 4, or compared to SRN in Stratum 3. Oversampling of subjects with adenocarcinoma of the colon or rectum is necessary to provide sufficient dedicated samples and data for validation trials aimed at regulatory approval. Samples and data from subjects recruited in this trial may be used to update and enhance reference sets used by the EDRN to further train, test and/or validate new biomarkers for future inclusion in validation trials aimed at regulatory approval. Strata 1-3 are necessary to ensure these comparison groups collected under the same protocol to Stratum 4 are available.
Training and validation: The prospective GLNE010 study has recruited many subjects in each stratum except adenocarcinoma. The samples from this protocol will need to be combined with GLNE010 samples to allow both panel building (training) and panel validation for each of above comparisons.
16
Assessment of the utility of individual biomarkers for discriminating between patients with adenocarcinomas, patients with adenomas, patients without adenomas and normal subjects.
For markers measured on a continuous scale, the within-class distributions of the marker values will be assessed by graphical means (e.g., q-q plots). Maximum likelihood estimates of distribution parameters will be calculated. For markers measured on a dichotomous scale, the proportions of positive tests in each class will be determined. For each marker, non-parametric (via SAS PROC LOGISTIC) and fully parametric ROC curves will be constructed for: Stratum 4 versus all others except SRN-adenomas (define as adenoma ≥1 cm or adenoma with high grade dysplasia or sessile serrated polyp ≥1 cm) (primary comparison); Stratum 4 versus Strata 1 and 2 (secondary comparison); and other exploratory comparisons: Stratum 4 versus Stratum 1; Stratum 4 and SRN-adenomas in Strata 3 versus Strata 1 and 2 and non-SRN adenoma; Strata 2, 3 and 4 versus Stratum 1. While the non-parametric ROCs are generally preferred, decision rules for population screens may require very high specificity, which will require accurate estimation in the distribution tails; parametric ROC curves may be better for this application. The area under each ROC curve (AUC) for each comparison will be determined.
Construction or testing of a panel of markers from those considered in Objective 1
Construction of a panel of markers from those considered in Objectives 1 and 2 to discriminate, under specific of assumptions concerning prevalence and cost of misclassification, for the primary, secondary, and exploratory comparisons described above. Candidate markers will be chosen according to both statistical (e.g., high patient or tissue sensitivity) and practical (less expensive assays, all markers assessed on blood) criteria. Forward stepwise logistic regression will be used to construct a panel to discriminate between the two classes of patients. The ROC curve will be constructed and AUC will be determined. Other panel building approach will also be used when appropriate, e.g. an “OR” rule will be used, that a test is positive if either one test is positive, if each of the biomarker is very specific but only for a subset of cancers.
Validation of a panel of biomarkers: If the cutoff has not been locked-down but the combination rule has been pre-determined, the optimal cutoff will depend on the intended clinical use. For example, for a blood based biomarker as a first step screening for those who do not want to do stool FIT tests, we might the cutoff that is corresponding to sensitivity of FIT test for colorectal cancers, then evaluate if the specificity of this cutoff is adequate. False positive is of less concern because it will lead to colonoscopy, a recommended screening in US. For a stool- based test (only for adenocarcinomas in this study when combined with the previous GLNE007 or GLNE010 set where stool samples were collected for all participants), if a test with much lower cost than that of the Exact Sciences multi-marker panel that includes a fecal immunochemical test and methylated DNA gene markers is brought for testing, the threshold sensitivity required to enter a large validation trial might be lower than that of the Exact Sciences panel (in the range of 85% to 92% for detection of adenocarcinoma) but better than FIT alone.
We would then compare the specificity to that of Exact Sciences multi-marker panel. If the cutoff has been pre-determined, then the evaluation will be a simple joint 1-sided 95% confidence region for sensitivity and specificity.
Comparison of the characteristics of individual markers and panels as discriminators to those of the established current standard, Fecal Immunohistochemistry test (FIT).
For biomarker validation we assume at least the panel combination rule has been locked-down. The biomarkers or panels could be from outside of the consortium or from the one built in 9.2.2 using previous GLNE007 specimens collected between 2006-2010 and GLNE 010 samples collected between 2011 and 2019. As in Objective 3, the following analysis will be performed for each of the primary, secondary, and exploratory comparisons. For blood-based biomarkers, we will test whether it has a similar sensitivity as that of FIT and has a reasonable specificity (e.g. > 70%) if the cutoff is pre-determined, or whether at a cutoff corresponding to the same sensitivity of FIT the specificity is better than 50% (target specificity > 70%) if a cutoff is not pre- determined. This performance criterion is also used for training set panel building, i.e., a panel will need to have this performance before it is locked-down for validation. For stool-based biomarkers, we will test whether the sensitivity is better than that of FIT, with compatible specificity. With non-screening colonoscopies, we will collect information whether the colonoscopy was triggered by a positive FIT test or triggered by symptom. The performance of biomarker will be evaluated with each of these two groups separately and compared, to gauge the potential bias caused by FIT positive results triggering colonoscopy that will lead to over- estimate of sensitivity for FIT.
17
Development of a bank of stool samples linked to serum, tissue, and clinical data from patients with colorectal cancer, adenomas and normal controls for validation of stool-based markers that may be developed in the future.
Markers available in the future will be developed in a similar fashion to Objectives 1-4. Every effort will be made to ensure that samples from and data concerning subjects in all four strata are collected, processed and stored according to the same procedures (Section 8.2 and Appendices), so that data and sample banking do not introduce bias into future studies.
9.3Justification of Design and Sample Size
The primary goals of this protocol are to enhance the already available EDRN reference set and provide biosamples and data as required to fill in validation sets for the purposes of regulatory approval. The reference sets and other GLNE 007 samples will be used to assess the ability of different markers to discriminate between patients with adenocarcinoma, patients with adenomas and normal subjects (Objective 1) and to strategically use this information to construct panels of markers to discriminate cases (adenocarcinomas and/or screen relevant-adenomas) from controls (Objective 2). An additional reference set might be set aside for the purposes of regulatory validation. Such samples may not be used for training or testing of a given marker that might be validated with samples from the GLNE 007 reference set or other validation sets previously collected by EDRN.
We justify the sample size for the primary comparison for training and validation separately:
Panel training and testing: Cancer (n=200) versus 560 normal (normal colonoscopies (200 average risk subjects, 200 high risk subjects) or non-screen relevant adenomas (estimated to be 80% of 200 adenomas, i.e. n=160)) for a blood-based test as the first line test for people who do not do any colorectal cancer screening. We assume the cutoff has not been locked-down (statistical power would be much larger if the cutoff is locked-down) so we will use cutoff corresponding to sensitivity of FIT (75%). We argue that with this sensitivity a test with at least 70% specificity would have great clinical utility. With the study sample size, we will have >90% power to reject a null hypothesis specificity of 58% if the true specificity is at least 70%.
For stratified analysis if there is evidence that specificities for normal colonoscopy high risk subjects and subjects with non-screen relevant adenoma are significantly lower than that for normal colonoscopy low risk group, suggesting they may have higher risk for screen relevant neoplasms in the future and should be analyzed separately. With 200 cancers and 200 normal colonoscopies in low risk group, we have at least 82% power to reject a null hypothesis specificity of 58% if the true specificity is at least 70%. With 200 cancers and 360 subjects in high risk or non-SRN adenoma groups, we have at least 89% power to reject a null hypothesis specificity of 58% if the true specificity is at least 70%.
For validation of stool-based test, we use the scenario that using a cutoff corresponding to 92% sensitivity (that of the Exact Sciences multi-marker panel) and test the adequacy of specificity, with 200 cancers and 747 normal or non-screen relevant neoplasms from GLNE010 training set, we have at least 86% power to reject a null hypothesis of specificity 76% if the true specificity is at least 85%. Such a test if it is substantially cheaper than that of the Exact Sciences multi-marker panel will have clinical value to increase the sensitivity of FIT.
Panel validation: Cancer (n=200) versus 560 normal (normal colonoscopies (200 average risk subjects, 200 high risk subjects) or non-screen relevant adenomas (estimated to be 80% of 200 adenomas, i.e. n=160)) for a blood-based test as the first line test for people who do not do any colorectal cancer screening. We assume the cutoff has not been locked-down (statistical power would be much larger if the cutoff is locked-down) so we will use cutoff corresponding to sensitivity of FIT (75%). We argue that with this sensitivity a test with at least 70% specificity would have great clinical utility. With the study sample size, we will have >90% power to reject a null hypothesis specificity of 58% if the true specificity is at least 70%. If the validation is done using all 3070 normal and non-screen relevant neoplasms from GLNE010 (the number as of April 2019), then we have >90% power for to reject a null hypothesis specificity of 60%.
For validation of stool-based test, we use the scenario that using a cutoff corresponding to 92% sensitivity (that of the Exact Sciences multi-marker panel) and test the adequacy of specificity, with 200 cancers from this protocol and 3070 normal and non-screen relevant neoplasms from GLNE010 as of April 2019, we have at least 90% power to reject a null hypothesis of specificity 76% if the true specificity is at least 85%. Such a test if it is substantially cheaper than that of the Exact Sciences multi-marker panel will have clinical value to increase the sensitivity of FIT.
18
10.0DATA SAFETY AND MONITORING
10.1Data Safety and Monitoring
Authority
The DSMC reviews, makes recommendations, and acts on the following:
a.All protocols being run through the GLNE EDRN will be monitored by the DSMC.
b.Progress towards completion of the trial—recruitment and retention of study participants.
c.Insufficient accrual to warrant continuation of the trial.
d.Evaluation of interim data analyses.
e.Evaluation of interim new information.
f.Evaluation of toxicity events including reporting of adverse events.
g.Timeliness of data.
h.Quality of data.
i.Ethical conduct of research.
The DSMC is empowered with the authority to recommend a trial be suspended or terminated based upon concerns in any of the above areas of review. The DSMC reviews all serious adverse events and ensures that these events have been correctly reported to all institutional review boards, and that adverse events have been correctly classified as serious or not serious. The Board assesses the impact of these events upon the conduct of the clinical trial. The Board is empowered with the authority to suspend or terminate any trials for which there are concerns of toxicity that endanger human participants. Monitoring also considers factors external to the study, such as scientific or therapeutic developments that may have an impact on the safety of the participants or the ethics of the study. Recommendations that emanate from monitoring activities are reviewed by the principal investigator and addressed.
Composition
The principal investigator is present in an open session portion of the meeting and absent in a closed session. All DSMC official subjects in the review of confidential data and discussions regarding continuance or stoppage of a study have no conflict of interest and no financial stake in the research outcome. The current UM Prevention research base Data and Safety Monitoring Committee is Chaired by the Research Base Biostatistician and comprised of Faculty members from Gastroenterology, Family Medicine, Hematology/Oncology. At least 3 faculty members, not including the study PI, must be present along with the biostatistician as chair to have quorum. If the DSMC cannot meet face-to-face, a conference call is acceptable.
Meeting Frequency
The UM Prevention Research Base DSMC meets monthly by means of regularly scheduled meetings. Prior to each meeting, the UM Prevention Research Base clinical research associate distributes a standard summary report detailing accrual, biomarker modulations data, new publications or presentations relevant to the ongoing project, quality control audit information, any ethical concerns, patient-subject complaints and adverse events or serious adverse events of all prevention protocols.
Recommendations and Reporting
Recommendations for action are sent to the Principal Investigator. The Principal Investigator is responsible for implementing DSMC recommendations. In addition to the Principal Investigator, minutes from the monthly meetings are forwarded to the following as needed:
a.DSMB members and the principal investigators at other sites
b.The University of Michigan Comprehensive Cancer Center Prevention and Control Protocol Review Committee Chair, per PRC policies;
c.IRBMED (University of Michigan Medical School IRB);
d.NCI/DCP Program Staff;
e.Any other trial sponsor.
Serious adverse events and adverse events are reported to the institutional review boards of all clinical sites, University of Michigan IRBMED per standard SAE reporting guidelines, and the sponsor as required by Federal regulations. A yearly summary report of trial activities is made to all trial investigators, supervisory committees and the sponsor. The UM prevention data management office and the DMCC have the responsibility of informing other trial investigators concerning the data and safety monitoring policy, procedures, and decisions.
19
Definition
An adverse event (AE) is any condition, which appears or worsens after the participant is enrolled in an investigational study.
AE Information
No adverse events are expected, as there is no intervention for this trial. Any adverse events related to the subject’s participation in this study will be forwarded to the data coordinating center and reported to the UM IRBMed per Standard Adverse Event Guidelines.
Serious Adverse Events
One-third of the participants will have colon cancer by study design, and deaths due to disease progression or serious adverse events due to cancer treatment are expected. The only procedures that are part of this study are blood, and stool collection, so it is unlikely that any deaths or hospitalizations will be related to the sample collection in this study. Only Serious Adverse Events that are deemed to be directly related to a study procedure (sample collection) by the DSMB will be reported to any regulatory body.
A serious adverse event is defined (by ICH Guideline E2A and Fed. Reg. 62, Oct. 7, 1997) as an event, occurring at any dose, which meets any of the following criteria:
Results in death
Is immediately life threatening
Requires inpatient hospitalization or prolongation of existing hospitalization
Results in persistent or significant disability/incapacity
Is a congenital anomaly/birth defect
In addition, events that may not meet these criteria, but which the investigator finds very unusual and/or potentially serious, will be reported in the same manner.
12.0DATA MANAGEMENT
12.1Registration
Institutional collaborators will enter IRB information into the secure VSIMS database, including IRB approval date, expiration date, and document versions. Subject registrations will not be allowed without IRB approval. The DMCC will provide recruiting sites with Participant ID numbers to be assigned in VSIMS. No exceptions to eligibility requirements will be permitted without prior permission of the protocol PI.
12.2Timeliness
Timeliness is monitored by the DMCC and UM through various reporting mechanisms within VSIMS.
12.3Completeness and Accuracy
The DMCC will assure the completeness of the data by writing data entry programs that will not allow for empty fields whenever possible. The accuracy of the data will be checked by identifying appropriate parameters allowed to be entered in a given data field. Periodic reviews of the paper CDEs and the database data will be conducted by the lead CRA and DMCC site monitor.
12.4Accuracy--Revisions and Corrections
All corrections to paper study documents will be initialed and dated. If computer-readable data is corrected by replacement of a data set, the replaced version of the data set will be retained in an archive. The collection of these auxiliary data sets represents an audit trail of corrections to the database.
20
12.5On Site Data Audits
All consortium sites are subject to periodic on-site audits. The objective of the on-site audit is to conduct a general review of a random sample of registered subjects from the selected protocol to assess overall protocol adherence with respect to subject eligibility, appropriate procedure for informed consent, registration process, general protocol adherence, sample shipment process, follow-up and off-study process.
An On-Site Audit checklist will be developed which will contain all of the essential elements of an On- Site audit. Each of the essential elements are reviewed and discussed with the clinical site. The Checklist is signed by the auditors and retained at the DMCC.
In preparation for a site audit, the study statistician will select the subjects for review using a randomized selection procedure. Other cases may also be selected at the discretion of the audit team. A minimum of 10% of the subjects accrued since the last audit will be reviewed. The on-site audit team will audit two to three unannounced cases. The consortium site investigator and research coordinator will be notified of the impending audit not more than 3 months in advance. Two to four weeks prior to the site visit, the list of selected subjects will be sent to the consortium clinical site. All data and material pertinent to the subject will be reviewed including eligibility criteria, informed consent, and sample shipment logs.
Subject data will be extracted at the DMCC prior to the visit. At the audit, the data from the DMCC will be compared to the original data (source documents and/or CDEs). On-site audit staff will review the documentation of IRB approvals, for each audited protocol, any amendments or adverse events, and consent forms.
Based on the findings of the audit, a follow-up schedule will be defined. A report of the audit is written and faxed to the DMCC and the NCI within 5 working days of the audit. A copy of the report is emailed or faxed to the consortium site investigator. The site PI has 30 days from receipt of the report to respond in writing to the DMCC directly. After the 30-day response period, the report is finalized and sent to NCI and the consortium site investigator.
The DMCC will maintain a file containing the latest version of the On-Site Audit guidelines, a listing of all consortium institutions reviewed to date, a copy of the On-Site Audit results and all correspondence for each audit conducted. These results will be reviewed by the Center’s Executive Committee at a monthly telephone conference and will be made available to the NCI.
12.6Sample Tracking
Sites receiving shipments of samples are notified via e-mail, so if samples are delayed or lost, tracking may be initiated by the sending site. Sample shipment forms are included with shipments. These data forms describe the date of sample receipt, and availability of sample, along with tracking information. The receiving site will evaluate the sample condition on arrival, scan the bar-coded samples in the VSIMS database, verify samples shipped match samples sent, and store at appropriate conditions until shipment to analytical labs.
12.7Confidentiality
Subjects will be identified in the database by their unique EDRN subject identification numbers only. Information that could identify subjects, such as name, address, or social security number will be kept only by the enrolling site and will not be supplied to the DMCC at Xxxx Hutch. The Coordinating Center at UM will have a separate payment form with name, address, and social security number for payment purposes only as previously described. During an on-site audit or NCI site visit, staff may review medical records and other information that contains PHI, but this information will not be removed from the enrolling site. The Coordinating center at UM will not keep copies of signed informed consent documents. No information, including copies of the informed consent unless required by the institution, obtained during the study will be placed in a subject’s medical record.
12.8Security
All subject files will be stored under lock and key at all times. All computer systems will be password- protected against intrusion; all network-based communications between sites of confidential information are encrypted.
An on-going computer-virus-protection program is available and used, maintained, and audited on all computers and pathways into the system, including good practice policies, screening of data files, executable software, diskettes, text macros, downloads, and other concerns as they arise. The DMCC will assist in maintaining appropriate levels of network security.
21
13.0ETHICAL AND REGULATORY CONSIDERATIONS
13.1Institutional Review
This study must be approved by an appropriate institutional review committee as defined by Federal Regulatory Guidelines (Ref. Federal Register Vol. 46, No. 17, January 27, 1981, part 56). The protocol and informed consent form for this study must be approved in writing by the appropriate Institutional Review Board (IRB). The IRB must be from an institution that has a valid Federal Wide Assurance, Multiple Project Assurance, Single Project Assurance or Cooperative Oncology Group Assurance on file with the Office for Human Research Protections, Department of Health and Human Services. The institution must comply with regulations of the Food and Drug Administration and the Department of Health and Human Services. Changes to the protocol, consent, as well as a changes to the investigator list at each site, must also be approved by the IRB and documentation of this approval provided to the Coordinating center. Records of the Institutional Review Board review and approval of all documents pertaining to this study must be kept on file by the investigator and are subject to OHRP or NCI inspection at any time during the study. Periodic status reports must be submitted to the Institutional Review Board at least yearly, as well as notification of completion of the study and a final report within 3 months of study completion or termination. The investigator must maintain an accurate and complete record of all submissions made to the Institutional Review Board, including a list of all reports and documents submitted.
Inclusion of New Biomarkers Discovered by EDRN Investigators over the Next Two Years
The design of this project including the collection of serum, DNA and tissue samples permit the inclusion of new EDRN discovered biomarkers into this panel. Should EDRN investigators provide sufficient preliminary data to justify inclusion in this panel; new biomarkers will be included in the validation program using the procedures described above.
22
14.0REFERENCES
1Lee JK, Xxxxx EG, Bent S, Xxxxx XX, Xxxxxx DA. Accuracy of fecal immunochemical tests for colorectal cancer: systematic review and meta-analysis. Xxx Intern Med. 2014;160(3):171.
2.Xxxxxxxxx XX, Xxxxxxxxx XX, Xxxxxxxxx XX, Xxxxx XX, Xxxxx P, Xxxxxxx XX, et al. Multitarget Stool DNA Testing for Colorectal-Cancer Screening. N Engl J Med. 2014.
3.Definitions Working Group, editor Biomarkers and Surrogate Endpoints. Biomarkers and Surrogate Endpoints; 1999; Bethesda, MD: National Institutes of Health, Food and Drug Administration.
4.Shatzkin A, Xxxxxxxx L, Xxxxxxxxx M, Sawsey S. Validation of intermediate end points in cancer research. J Natl Cancer Inst. 1990;82:1746-52.
5.Xxxxxxxx XX. Surrogate endpoints in clinical trials: definition and operational criteria. Stat Med. 1989;8(4):431-40.
6.Xxxxxxx S, Lec J, Lotan R, Hittelman W, Xxxxxxxxx M, Hong W. Biomarkers as intermediate endpoints in chemoprevention trials. JNCI. 1990;82:555-60.
7.Xxxxxx XX, Xxxx XX, Church TR, Xxxxxx XX, Xxxxxxx XX, Xxxxxxx XX, et al. Reducing mortality from colorectal cancer by screening for fecal occult blood. Minnesota Colon Cancer Control Study [published erratum appears in N Engl J Med 1993 Aug 26;329(9):672] [see comments]. N Engl J Med. 1993;328(19):1365-71.
8.Kronborg O, Xxxxxx C, Xxxxx J, Xxxxxxxxx O, Xxxxxxxxxxx O. Randomised study of screening for colorectal cancer with faecal-occult-blood test. Lancet. 1996;348:1467-71.
9.Xxxxxxxxxx XX, Xxxxxxxxxxx XX, Xxxxxxxx XX, Xxxx XX, Amar SS, Xxxxxxx XX, et al. Randomised controlled trial of faecal-occult-blood screening for colorectal cancer. Lancet. 1996;348(9040):1472-7.
10.Xxxxxxx A, Xxxxxx XX, Xxxxxxx MS, Xxxxxxx XX, Xxxx XX, Xxxxxx XX, et al. Long-term mortality after screening for colorectal cancer. N Engl J Med. 2013;369(12):1106-14.
11.Xxxxxx NN, Xxxxxxxxxx MA, Paszat LF, Saskin R, Xxxxxx DR, Xxxxxxxx L. Association of colonoscopy and death from colorectal cancer. Xxx Intern Med. 2009;150(1):1-8.
12.Singh H, Xxxxxx Z, Xxxxxx XX, Xxxxxxx XX, Xxxxxx XX, Xxxxxxxxx CN. The reduction in colorectal cancer mortality after colonoscopy varies by site of the cancer. Gastroenterology. 2010;139(4):1128-37.
13.Xxxxxxx H, Xxxxx-Xxxxxx J, Xxxxxx XX, Xxxxxxx A, Xxxxxxxxxxx M. Protection from colorectal cancer after colonoscopy: a population-based, case-control study. Xxx Intern Med. 2011;154(1):22-30.
14.Muto T, Kamiya J, Sawada T, Konishi F, Sugihara K, Kubota Y, et al. Small flat adenoma of the large bowel with special reference to its clinicopathologic features. Dis Colon Rectum. 1985;28(11):847-51.
15.Rex DK, Xxxxxx XX, Xxxxxx XX, Xxxxxxx XX, Xxxxx XX, Helper DJ, et al. Colonoscopic miss rates of adenomas determined by back-to-back colonoscopies. Gastroenterology. 1997;112(1):24- 8.
16.Saitoh Y, Waxman I, West AB, Xxxxxxxxxx XX, Xxxxxxxx Z, Watari J, et al. Prevalence and distinctive biologic features of flat colorectal adenomas in a North American population. Gastroenterology. 2001;120(7):1657-65.
17.Xxxxxxxx NJ, Xxxxxx K, Xxxxx XX, Xxxxxx L, Xxxxxxx K, Xxxxxxxxxx D, et al. Characteristics of missed or interval colorectal cancer and patient survival: a population-based study. Gastroenterology. 2014;146(4):950-60.
18.Xxxxxxx XX, Xxxxxx XX, Xxxxxxxx XX. Effect of a time-dependent colonoscopic withdrawal protocol on adenoma detection during screening colonoscopy. Clin Gastroenterol Hepatol. 2008;6(10):1091-8.
19.Xxxxxxx XX, Xxxxxx XX, Xxxxxxx AS, Xxxxxxxx XX, Xxxxxxxx XX. Colonoscopic withdrawal times and adenoma detection during screening colonoscopy. N Engl J Med. 2006;355(24):2533-41.
00.Xxxxxxxx XX, Xxxxxxxxxx T, Xxxxx XX, Park W, Xxxxxxxxxx A, Sato T, et al. Prevalence of nonpolypoid (flat and depressed) colorectal neoplasms in asymptomatic and symptomatic adults. JAMA. 2008;299(9):1027-35.
21.Zauber AG, Xxxxxxx XX, O’Xxxxx XX, Xxxxxxxx-Xxxxxxxx I, van Ballegooijen M, Xxxxxx XX, et al. Colonoscopic polypectomy and long-term prevention of colorectal-cancer deaths. N Engl J Med. 2012;366(8):687-96.
22.Xxxxx D, Vanaclocha M, Ibanez J, Xxxxxx-Xxxxxxx A, Xxxxxxxxx V, Cubiella J, et al. Participation and detection rates by age and sex for colonoscopy versus fecal immunochemical testing in colorectal cancer screening. Cancer Causes Control. 2014;25(8):985-97.
23.Xxxxxxxx E, Castells A, Xxxxxxx L, Cubiella J, Xxxxx D, Lanas A, et al. Colonoscopy versus fecal immunochemical testing in colorectal-cancer screening. N Engl J Med. 2012;366(8):697-706.
24.Xxxxx XX, Xxxxx XX. Overdiagnosis in cancer. J Natl Cancer Inst. 2010;102(9):605-13.
25.Xxxxx B, Xxxxxxxxx DA, XxXxxxxxx B, Xxxxxxx KS, Xxxxxx D, Bond J, et al. Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: a joint guideline from the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. Gastroenterology. 2008;134(5):1570-95.
26.Force USPST. Screening for colorectal cancer: U.S. Preventive Services Task Force recommendation statement.[see comment][summary for patients in Xxx Intern Med. 2008 Nov 4;149(9):I-44; PMID: 18838719]. Annals of Internal Medicine. 2008;149(9):627-37.
00.Xxxxxxxxx NC. International Cancer Screening Network Xxxxxxxx, XX0000 [Available from: xxxx://xxxxxxxxxxxxxxx.xxxxxx.xxx/xxxx/xxxxxxxxxx/xxxxxxxxx.xxxx.
23
28.Pox CP, Xxxxxxxxxx L, Xxxxxxx H, Theilmeier A, Von Stillfried D, Schmiegel W. Efficacy of a nationwide screening colonoscopy program for colorectal cancer. Gastroenterology. 2012;142(7):1460-7 e2.
29.Centers for Disease C, Prevention. Xxxxx xxxxx: colorectal cancer screening test use--United States, 2012. MMWR Morb Mortal Wkly Rep. 2013;62(44):881-8.
30.Surveillance E, and End Results Program. SEER Stat Fact Sheets: Colon and Rectum Cancer: Centers for Disease Control; 2014 [Available from: xxxx://xxxx.xxxxxx.xxx/xxxxxxxxx/xxxx/xxxxxxxx.xxxx.
00.Xxxxx BB, Xxxx XX, Xxxxxxxx XX, Xxxxxx J, Xxxxxx XX, Xxxxxx XX, et al. An automated intervention with stepped increases in support to increase uptake of colorectal cancer screening: a randomized trial. Xxx Intern Med. 2013;158(5 Pt 1):301-11.
32.Xxxxxxxx XX, Xxxxxxxx XX, Xxxxxx XX, 3rd. A prospective, controlled assessment of factors influencing acceptance of screening colonoscopy. Am J Gastroenterol. 2002;97(12):3186-94.
33.Xxxxxxxxxx C, Xxxxxx M, Xxxxx XX, Xxxxx I, Xxxx Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra24.
34.Hibi K, Xxxxxxxx XX, Xxxxxx S, Wu L, Xxxxxxxx SR, Sidransky D, et al. Molecular detection of genetic alterations in the serum of colorectal cancer patients. Cancer Res. 1998;58(7):1405-7.
35.Vlems FA, Xxxxxxxx XX, Cornelissen IM, Xxxxxxxxxx XX, Wobbes T, Xxxx XX, et al. Investigations for a multi-marker RT-PCR to improve sensitivity of disseminated tumor cell detection. Anticancer Res. 2003;23(1A):179-86.
36.Yamaguchi K, Takagi Y, Aoki S, Futamura M, Xxxx X. Significant detection of circulating cancer cells in the blood by reverse transcriptase-polymerase chain reaction during colorectal cancer resection. Xxx Surg. 2000;232(1):58-65.
37.Zou H, Yu X, Xxxx X, Xxx X, Xxxx X, Xxx F, et al. Detection of aberrant p16 methylation in the serum of colorectal cancer patients. Clin Cancer Res. 2002;8:188-91.
38.Noh YH, Im G, Xx XX, Xxx XX, Xxx XX. Detection of tumor cell contamination in peripheral blood by RT-PCR in gastrointestinal cancer patients. J Korean Med Sci. 1999;14(6):623-8.
39.Xxxxx XX, Rajput A, Xxxxxxxxxxx XX, Xxxxxxxxx SD. Detection of aberrantly methylated hMLH1 promoter DNA in the serum of patients with microsatellite unstable colon cancer. Cancer Res. 2001;61(3):900-2.
40.Nakayama H, Hibi K, Taguchi M, Takase T, Yamazaki T, Kasai Y, et al. Molecular detection of p16 promoter methylation in the serum of colorectal cancer patients. Cancer Lett. 2002;188(1- 2):115-9.
41.Verma M, Srivastava S. Epigenetics in cancer: implications for early detection and prevention. Lancet Oncol. 2002;3(12):755-63.
00.Xx M, Xxxx XX, Xxxxxxxxxxxx N, Xxxxxxx XX, Xxxxxxxxxxx NC, Xxxxxxxx S, et al. Sensitive digital quantification of DNA methylation in clinical samples. Nat Biotechnol. 2009;27(9):858- 63.
43.Grutzmann R, Xxxxxx B, Pilarsky C, Xxxxxxxxx XX, Xxxxxx PM, Xxxxxx HD, et al. Sensitive detection of colorectal cancer in peripheral blood by septin 9 DNA methylation assay. PLoS One. 2008;3(11):e3759.
44.Xxxxxx-Day C, Model F, Xxxxx T, Xxxxxxx R, Xxxxxxx J, Xxxxxxxx M, et al. DNA methylation biomarkers for blood-based colorectal cancer screening. Clin Chem. 2008;54(2):414-23.
00.Xxxxxx TR, Xxxxxxx M, Xxxxxx-Day C, Xxxxxx XX, Burger M, Xxxxx SR, et al. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut. 2014;63(2):317-25.
46.Hofsli E, Xxxxxxx W, Prestvik WS, Xxxxxxxx J, Rye M, Trano G, et al. Identification of serum microRNA profiles in colon cancer. Br J Cancer. 2013;108(8):1712-9.
47.Apweiler R, Hermjakob H, Xxxxxx X. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta. 1999;1473(1):4-8.
48.Xxxxxxxx B, Tharmalingam T, Xxxx PM. Glycans as cancer biomarkers. Biochim Biophys Acta. 2012;1820(9):1347-53.
49.Xxxxx TH, Zhao J, Xxxxxxxx MA, Xxxxxxx XX, Xxxxxx XX. Screening of glycosylation patterns in serum using natural glycoprotein microarrays and multi-lectin fluorescence detection. Anal Chem. 2006;78(18):6411-21.
50.Yue T, Xxxxxx XX, Xxxxxx B, Li L, Xxxxxxx K, Xxxxxxxx MA, et al. Enhanced discrimination of malignant from benign pancreatic disease by measuring the CA 19-9 antigen on specific protein carriers. PLoS One. 2011;6(12):e29180.
51.Zhao J, Xxxxx TH, Qiu W, Xxxxxxx K, Xxxxxxxx R, Xxxxx DE, et al. Glycoprotein microarrays with multi-lectin detection: unique lectin binding patterns as a tool for classifying normal, chronic pancreatitis and pancreatic cancer sera. J Proteome Res. 2007;6(5):1864-74.
52.Zhao J, Qiu W, Xxxxxxx XX, Xxxxxx XX. N-linked glycosylation profiling of pancreatic cancer serum using capillary liquid phase separation coupled with mass spectrometric analysis. J Proteome Res. 2007;6(3):1126-38.
53.Qiu Y, Xxxxx TH, Xu L, Xxxxxxx K, Xxxxx DE, Xxxx M, et al. Plasma glycoprotein profiling for colorectal cancer biomarker identification by lectin glycoarray and lectin blot. J Proteome Res. 2008;7(4):1693-703.
54.Rho JH, Xxxx JR, Xxxxxx XX, Xxxxxxx DE, Xxxxx XX, Xxxxxxxxxxxx XX, et al. Discovery of sialyl Xxxxx A and Xxxxx X modified protein cancer biomarkers using high density antibody arrays. J Proteomics. 2014;96:291-9.
55.Xxxxxxxxx XX, Xxxx XX, Xxxxxxx D, Xxxxx J, Xxxxxx J, Xxxxx D, et al. A circulating ligand for galectin-3 is a haptoglobin-related glycoprotein elevated in individuals with colon cancer. Gastroenterology. 2004;127(3):741-8.
24
56.Xxxxxxx XX, Welt S, Xxxxxx XX, Chen YT, Gure AO, Xxxxxxxx E, et al. Cancer-related serological recognition of human colon cancer: identification of potential diagnostic and immunotherapeutic targets. Cancer Res. 2002;62(14):4041-7.
00.Xx H, Xxxxxxx V, Xxxxx XX. Targeting serum antibody for cancer diagnosis: a focus on colorectal cancer. Expert Opin Ther Targets. 2007;11(2):235-44.
58.Xxx XX, Madoz-Gurpide J, Wang H, Lescure P, Xxxxxxxxxx XX, Xxxx R, et al. Molecular profiling of the immune response in colon cancer using protein microarrays: occurrence of autoantibodies to ubiquitin C-terminal hydrolase L3. Proteomics. 2003;3(11):2108-15.
59.Zaenker P, Xxxxx MR. Serologic autoantibodies as diagnostic cancer biomarkers--a review. Cancer Epidemiol Biomarkers Prev. 2013;22(12):2161-81.
60.Xxxxxxx XX, Tanneberger S, Pannetta A, Bedosti M, Poell M, Xxxxxxxxxx K, et al. Increase in autoantibodies against Fas (CD95) during carcinogenesis in the human colon: a hope for the immunoprevention of cancer? Cancer Immunol Immunother. 2005;54(10):1038-42.
61.He Y, Wu Y, Mou Z, Li W, Zou L, Fu T, et al. Proteomics-based identification of HSP60 as a tumor-associated antigen in colorectal cancer. Proteomics Clin Appl. 2007;1(3):336-42.
62.Chen Y, Lin P, Qiu S, Peng XX, Looi K, Xxxxxxxx XX, et al. Autoantibodies to Ca2+ binding protein Calnuc is a potential marker in colon cancer detection. Int J Oncol. 2007;30(5):1137-44.
63.Liu W, Wang P, Li Z, Xu W, Dai L, Wang K, et al. Evaluation of tumour-associated antigen (TAA) miniarray in immunodiagnosis of colon cancer. Scand J Immunol. 2009;69(1):57-63.
64.Giovannucci E. Insulin-like growth factor-1 and binding protein-3 and risk of cancer. Horm Res. 1999;51(Suppl 3):34-41.
65.Giovannucci E, Xxxxxx MN, Xxxxx XX, Xxxxxx XX, Xxxxxxxx XX, Xxxxxx N, et al. A prospective study of plasma Insulin-like growth factor and binding protein-3 and risk of colorectal neoplasia in women. Cancer Epidemiol Biomarkers Prev. 2000;9(4):345-9.
66.Giovannucci E, Xxxxxx M, Xxxxx XX, Xxxxxx XX, Xxxxxxxx XX, Xxxxxx N, et al. Insulin-like growth factor I (IGF-I), IGF-binding protein-3 and the risk of colorectal adenoma and cancer in the Nurses’ Health Study. Growth Horm IGF Res. 2000;10 Suppl A:S30-1.
67.Xxxxxxxxx R, Stattin P, Xxxxxxx S, Biessy C, Stenling R, Riboli E, et al. Plasma insulin, IGF- binding proteins-1 and -2 and risk of colorectal cancer: a prospective study in northern Sweden. Int J Cancer. 2003;107(1):89-93.
68.Xxxx-Xxxxxx M, Cui H, Giardiello FM, Xxxx XX, Xxxxxx L, Xxxxxxxx A, et al. Loss of imprinting of insulin growth factor II gene: a potential heritable biomarker for colon neoplasia predisposition. Gastroenterology. 2004;126(4):964-70.
69.Cui H, Onyango P, Xxxxxxxxxxx S, Wu Y, Xxxxx XX, Xxxxxxxx XX. Loss of imprinting in colorectal cancer linked to hypomethylation of H19 and IGF2. Cancer Res. 2002;62(22):6442-6.
70.Cui H, Xxxx-Xxxxxx M, Giardiello FM, Xxxxxxxx XX, Kafonek DR, Xxxxxxxxxxx S, et al. Loss of IGF2 imprinting: a potential marker of colorectal cancer risk. Science. 2003;299(5613):1753-5.
71.Xxxxxxx K, Flood A, Green L, Xxxxxxx XX, Xxxxxx J, Cash B, et al. Loss of insulin-like growth factor-II imprinting and the presence of screen-detected colorectal adenomas in women. J Natl Cancer Inst. 2004;96(5):407-10.
72.Tsushima H, Kawata S, Tamura S, Ito N, Shirai Y, Kiso S, et al. High levels of transforming growth factor beta 1 in patients with colorectal cancer: association with disease progression. Gastroenterology. 1996;110(2):375-82.
73.Tsushima H, Ito N, Tamura S, Matsuda Y, Inada M, Yabuuchi I, et al. Circulating Transforming Growth Factor beta-1 as a predictor of liver metastasis after resection in colorectal cancer. Clin Cancer Res. 2001;7:1258-62.
74.Narai S, Watanabe M, Hasegawa H, Nishibori H, Endo T, Kubota T, et al. Significance of Transforming growth factor beta 1 as a new tumor marker for colorectal cancer. Int J Cancer. 2002;97(4):508-11.
75.Xxxxx R, Xxxxxxx H, Xxxxxxx M, Xxxxxxxxx E, Xxxxxxxxxx O, Xxxxxxx U, et al. Vascular endothelial growth factor (VEGF) -- a valuable serum tumour marker in patients with colorectal cancer? Eur J Surg Oncol. 2001;27(1):37-42.
76.Takeda A, Shimada H, Imaseki H, Okazumi S, Natsume T, Suzuki T, et al. Clinical significance of serum vascular endothelial growth factor in colorectal cancer patients : correlation with clinicopathological factors and tumor markers. Oncol Rep. 2000;7(2):333-8.
77.Shimoyama S, Xxxxxxxx K, Xxxxxxxx M, Kaminishi M. Increased serum angiogenin concentration in colorectal cancer is correlated with cancer progression. Clin Cancer Res. 1999;5(5):1125-30.
78.Xxxxxxx AL, Xxxxxxxxx XX, Jr, Xxxxxxxx XX, Xxxxxx XX, Xxxxxx MS, Xxxxxxxxx XX, et al. A prospective analysis of plasma endostatin levels in colorectal cancer patients with liver metastases. Xxx Surg Oncol. 2001;8(9):741-5.
79.Xxxxxxx XX, Xxxxxxxxx T, Xxxxxx XX, London NJ, Xxxxxxxxx XX. Raised levels of plasma big endothelin 1 in patients with colorectal cancer. Br J Surg. 2000;87(10):1409-13.
80.Xxxxxxx XX, Xxxxxx XX, Sweep FC, Span PN, Wobbes T, Ruers TM. Elevated serum endothelin-1 levels in patients with colorectal cancer; relevance for prognosis. Int J Biol Markers. 2000;15(4):288-93.
81.Xxxxxxxxxx P, Contasta I, Berghella AM, Xxxxxxx E, Xxxxxxxxxx C, Xxxxxx D. Simultaneous measurement of soluble carcinoembryonic antigen and the tissue inhibitor of metalloproteinase TIMP 1 serum levels for use as markers of pre-invasive to invasive colorectal cancer. Cancer Immunol Immunother. 2000;49(7):388-94.
25
82.Yukawa N, Yoshikawa T, Akaike M, Sugimasa Y, Takemiya S, Yanoma S, et al. Plasma concentration of tissue inhibitor of matrix metalloproteinase 1 in patients with colorectal carcinoma. Br J Surg. 2001;88(12):1596-601.
83.Barozzi C, Ravaioli M, X’Xxxxxx A, Xxxxx XX, Xxxxxxxx G, Xxxxxxx G, et al. Relevance of biologic markers in colorectal carcinoma: a comparative study of a broad panel. Cancer. 2002;94(3):647- 57.
84.Xxxxxx-Xxxxxxxx MN, Xxxxxxxxxxx XX, Xxxxxxx XX, Xxxxxxxx XX, Xxxxxx V, Xxxxxxx OH, et al. Total levels of tissue inhibitor of metalloproteinases 1 in plasma yield high diagnostic sensitivity and specificity in patients with colon cancer. Clin Cancer Res. 2002;8(1):156-64.
85.Alexiou D, Xxxxxxxxxxxxx XX, Xxxxxxx XX, Zbar A, Kremmyda A, Bramis I, et al. Serum levels of E-selectin, ICAM-1 and VCAM-1 in colorectal cancer patients: correlations with clinicopathological features, patient survival and tumour surgery. Eur J Cancer. 2001;37(18):2392-7.
86.Xxxxx XX, Xxxxxxx XX. Circulating tumor cells. Prog Mol Biol Transl Sci. 2010;95:95-112.
87.Wicha MS, Xxxxx XX. Circulating tumor cells: not all detected cells are bad and not all bad cells are detected. J Clin Oncol. 2011;29(12):1508-11.
88.Xxx XX, Xxxxxx XX, Chua W, Xx XX, xx Xxxxx P, Spring KJ. Circulating tumour cells and the epithelial mesenchymal transition in colorectal cancer. J Clin Pathol. 2014;67(10):848-53.
89.Zhang Z, Xxxxxxx X. Microfluidics and cancer: are we there yet? Biomed Microdevices. 2013;15(4):595-609.
90.Xxxxx XX, Xxx XX, Xxxxxxx S, Yu M, Xxxxxxxx XX, Ulkus L, et al. Isolation and characterization of circulating tumor cells from patients with localized and metastatic prostate cancer. Sci Transl Med. 2010;2(25):25ra3.
91.Xxxxxxxxx V, Xxxxxxx M, Grabauskiene S, Xxxxxxx-Xxxxxx M, Wicha MS, Xxxxxxx XX, et al. A radial flow microfluidic device for ultra-high-throughput affinity-based isolation of circulating tumor cells. Small. 2014;10(23):4895-904.
92.Xxxxxx XX. Galectinomics: finding themes in complexity. Biochim Biophys Acta. 2002;1572(2- 3):209-31.
93.Xxxxx XX, Xxxxxx XX, Xxxxxxxxx XX, Xxxx XX, Xxxxxxxxx XX. Expression of human intestinal mucin is modulated by the beta-galactoside binding protein galectin-3 in colon cancer. Gastroenterology. 2002;123(3):817-26.
94.Xxxxxxx N, Xxxxxxx J, Xxxx XX, Raz A, Xxxxxxxxx XX. Phosphorylation of the beta-galactoside- binding protein galectin-3 modulates binding to its ligands. J Biol Chem. 2000;275(46):36311-5.
95.Xxxxxxxxxx H, Raz A, Ho S, Xxxxxxxxx R. Expression of an endogenous galactose-binding lectin correlates with neoplastic progression in the colon. Cancer. 1995;75:2818-26.
96.Xxxxxxx X, Xxxxxxxxx XX, Castells A, Xxxxxxxxxx V, van den Brule F, Liu FT, et al. Differential expression of galectin 3 and galectin 1 in colorectal cancer progression. Gastroenterology. 1997;113(6):1906-15.
97.Xxxx XX, Xxxxxxx XX, Jr., Xxxxxxxxx CA, Xxx XX, Xxxxxx XX, Jr., Xxxxxx A, et al. Decreased expression of Mac-2 (carbohydrate binding protein 35) and loss of its nuclear localization are associated with the neoplastic progression of colon carcinoma. Proc Natl Acad Sci U S A. 1993;90(8):3466-70.
98.Levi Z, Rozen P, Hazazi R, Xxxxxx A, Waked A, Maoz E, et al. A quantitative immunochemical fecal occult blood test for colorectal neoplasia. Xxx Intern Med. 2007;146(4):244-55.
99.Shastri YM, Xxxxx J. Quantitative immunochemical fecal occult blood test for diagnosing colorectal neoplasia. Xxx Intern Med. 2007;147(7):522-3; author reply 3.
100.Rabeneck L, Rumble RB, Xxxxxxxx F, Xxxxx M, Xxxxxxxx C, Whibley A, et al. Fecal immunochemical tests compared with guaiac fecal occult blood tests for population-based colorectal cancer screening. Can J Gastroenterol. 2012;26(3):131-47.
101.Xxxxxxxx XX, Xxx XX, Xxxxx E, Xxxx XX, Fu R. Screening for colorectal cancer: a targeted, updated systematic review for the U.S. Preventive Services Task Force. Xxx Intern Med. 2008;149(9):638-58.
102.Xxxx XX, Pellise M, Xxxxxxxx XX, Xxxxxxxxx C, Xxxxxx M, Xxxx J, et al. Risk stratification for advanced colorectal neoplasia according to fecal hemoglobin concentration in a colorectal cancer screening program. Gastroenterology. 2014;147(3):628-36 e1.
103.Xxxxxxxx DA, Xxxxxx XX. Stool screening for colorectal cancer: evolution from occult blood to molecular markers. Clin Chim Acta. 2002;315(1-2):157-68.
104.Xxxxxx XX, Xxxxxxxx DA. Stool screening for colorectal cancer: molecular approaches. Gastroenterology. 2005;128(1):192-206.
105.Xxxxxxxx DA, Xxxxxxx XX, Xxxxxxxx XX, Xxxxx XX, Xxx XX, Xxxxx XX, et al. Stool DNA and occult blood testing for screen detection of colorectal neoplasia. Xxx Intern Med. 2008;149(7):441-50, W81.
106.Xxxxxxxxx XX, Xxxxxxxxx XX, Xxxxxxxxx XX, Xxxxxxxx XX, Xxxx ME. Fecal DNA versus fecal occult blood for colorectal-cancer screening in an average-risk population. N Engl J Med. 2004;351(26):2704-14.
107.Force USPST. Final Research Plan: Colorectal Cancer Screening 2014 [Available from: xxxx://xxx.xxxxxxxxxxxxxxxxxxxxxxxxxxxxx.xxx/Xxxx/Xxxxxxxx/XxxxxxxxXxxxXxxxx/xxxxxxxxxx- cancer-screening2.
108.Xxxx XX, Han ZJ, Skoletsky J, Xxxxx J, Xxx J, Xxxxxxx L, et al. Detection in fecal DNA of colon cancer-specific methylation of the nonexpressed vimentin gene. J Natl Cancer Inst. 2005;97(15):1124-32.
109.Zou H, Xxxxxxxxxx XX, Shire AM, Xxxx XX, Wang L, Xxxxxxxx ME, et al. Highly methylated genes in colorectal neoplasia: implications for screening. Cancer Epidemiol Biomarkers Prev. 2007;16(12):2686-96.
26
110.Xxxxxxxxx S, Brand R, Jandorf L, Xxxxxx K, Xxxxxxxxxxx J, Xxxxxxxx L, et al. A simplified, noninvasive stool DNA test for colorectal cancer detection. Am J Gastroenterol. 2008;103(11):2862-70.
111.Xxxxxxxxx XX, Jandorf L, Brand R, Rabeneck L, Xxxxxx PC, 3rd, Xxxxxx S, et al. Improved fecal DNA test for colorectal cancer screening. Clin Gastroenterol Hepatol. 2007;5(1):111-7.
112.Xxxxxxxx XX, Xxxxxx MA, Xxxxxx MTt, Xxxxxxx XX. The human gut microbiome as a screening tool for colorectal cancer. Cancer Prev Res (Phila). 2014;7(11):1112-21.
113.Xxxxxx AD, Xxxx X, Xxxxxxxxx X, Xxxxxxxx XX, Xxxxxxx XX, Xxxxxxx M, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14(2):207-15.
114.Montrose DC, Zhou XK, Kopelovich L, Xxxxxxx XX, Xxxxxx XX, Xxxxxxxxxxxx K, et al. Metabolic profiling, a noninvasive approach for the detection of experimental colorectal neoplasia. Cancer Prev Res (Phila). 2012;5(12):1358-67.
115.Ahmed FE, Ahmed NC, Vos PW, Bonnerup C, Xxxxxx XX, Xxxxx M, et al. Diagnostic microRNA markers to screen for sporadic human colon cancer in stool: I. Proof of principle. Cancer Genomics Proteomics. 2013;10(3):93-113.
000.Xxxx A, Xxxxxxxx F, Xxxx Y, Nagasaka T, Xxxxxx XX, Xxxxxx XX, et al. Fecal MicroRNAs as novel biomarkers for colon cancer screening. Cancer Epidemiol Biomarkers Prev. 2010;19(7):1766-74.
117.Vogelstein B, Xxxxxxxxxxxx N, Velculescu VE, Zhou S, Xxxx LA, Jr., Xxxxxxx XX. Cancer genome landscapes. Science. 2013;339(6127):1546-58.
118.Xxxxxxxxx DA, Xxxxxxxx XX, Xxxxx XX, Xxxxxx XX, Xxxxx XX, Xxxxx XX, et al. Race, ethnicity, and sex affect risk for polyps >9 mm in average-risk individuals. Gastroenterology. 2014;147(2):351-8; quiz e14–5.
119.Xxxxxxxxx DA, Xxxxx XX, Xxxxxx XX, Xxxxx J, Xxxxxxxx XX, Xxxxxx P. Low rate of large polyps (>9 mm) within 10 years after an adequate baseline colonoscopy with no polyps. Gastroenterology. 2014;147(2):343-50.
120.Xxxxxxxx XX, Xxxxxxxxx XX. Tailoring colonoscopic screening to individual risk. Gastroenterology. 2014;147(2):264-6.
121.Moinova H, Xxxxxxx XX, Xxxx L, Lutterbaugh J, Xxxxxxxxx-Xxxxx JS, Chen Y, et al. Aberrant vimentin methylation is characteristic of upper gastrointestinal pathologies. Cancer Epidemiol Biomarkers Prev. 2012;21(4):594-600.
27
EXHIBIT B
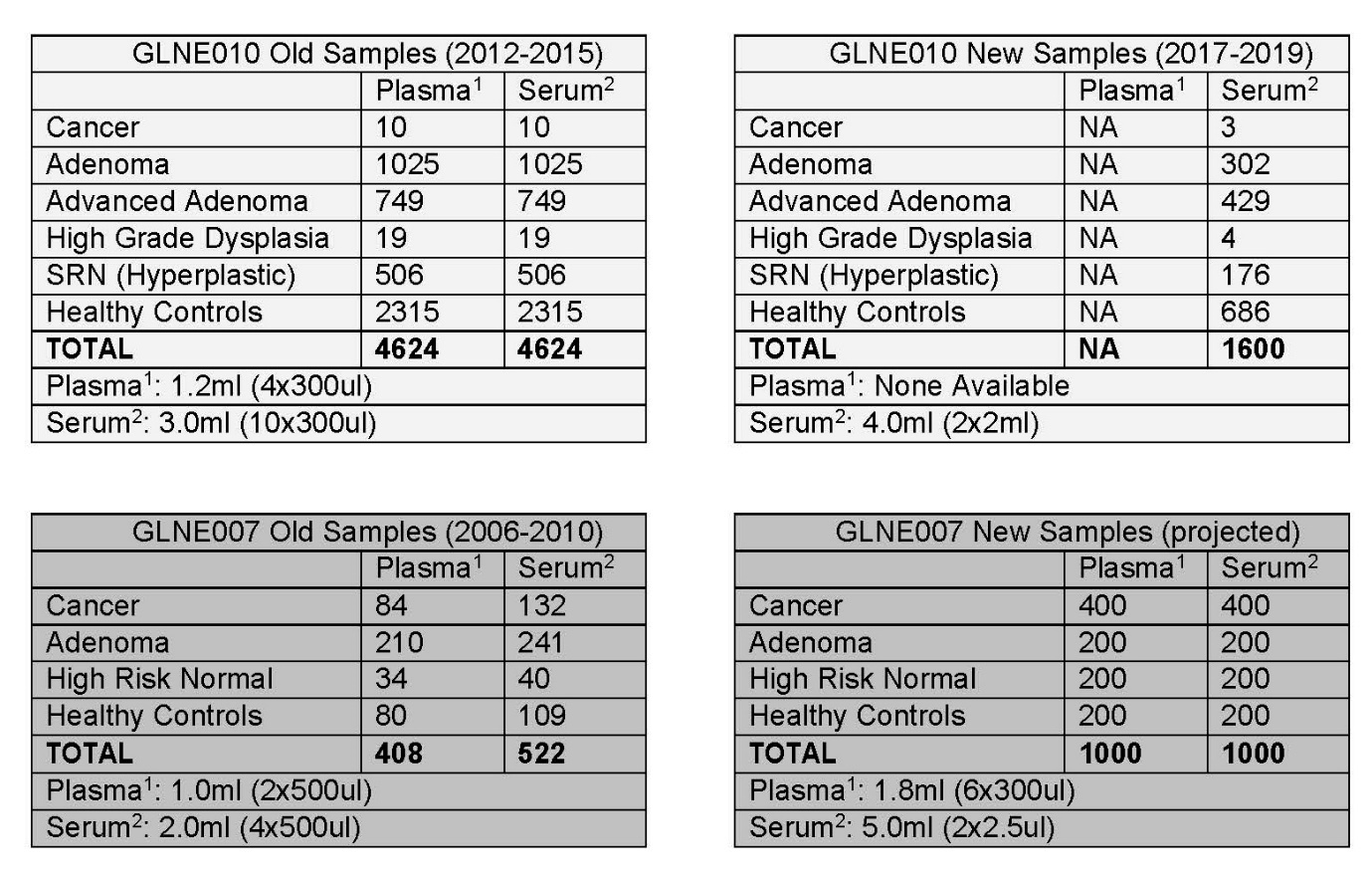
28
EXHIBIT C
Statement of Work (for University of Michigan Grant Activity) Volition
Aims
Aim 1: Complete GLNE 007, a trial designed to train and test and circulating biomarkers for early detection of colorectal adenocarcinoma.
Aim 2: To perform phase 1 validation trials (training and test set designs) of promising biomarkers discovered by EDRN Biomarker Validation Laboratories, external academic collaborating institutions, and collaborating EDRN industrial partners for the early detection of colorectal cancer.
Aim 3: To enhance and curate an archive of appropriately preserved stool, serum, plasma, urine, tissue and DNA biospecimens to be used by EDRN investigators for future validation and biomarker discovery research.
Work Plan
As part of the overarching EDRN project, adults age 50 or older undergoing a screening or surveillance colonoscopy will be enrolled as Study Subjects for this Project. Samples obtained from these Study Subjects will be sent to the GLNE Central Laboratory for preparation for storage/shipment to Volition (Laboratory). There, the samples will be tested for blood-based, cell-free circulating biomarkers on their proprietary Nu.QTM platform.
Laboratory will provide a copy of all test results for the Clinical Study to Principal Investigator’s designated Data Management Coordinating Center (DMCC) following the completion of their services. The DMCC will collect and store all such test results, and shall share test results with Institutions for collaborative analysis.
29
GLNE0l0
VALIDATION AND COMPARISON OF BIOMARKERS FOR THE EARLY
DETECTION OF COLORECTAL ADENOCARCINOMA
Great Lakes New England Clinical Validation Center
NCI Early Detection Research Network
2 UOl CA086400-16
Xxxx X. Xxxxxxx, M.D.1
Xxxx X. Xxxxx, M.D. 2
Xxxxxxx Xxxxxxx, M.O.3
Xxxxxx Xxxxxxxxx, M.D. 4
Xxx Xxxxxxx, M.D.6
Xxxxxxx Xxxxxx, PhD5
Xxxx Xxxxxxxx, M.O.2
Xxxx Xxxxxx, M.D. 7
Xxxxx Xxxxxx, M.D. 8
Ananda Sen, Ph.D.1
Ziding Feng, Ph.D.4
Xxxxxxxx Xxxx, Ph.D.9
Xxxxxxx Xxxx, M.S.1
1 University of Michigan Medical Center, Ann Arbor, MI
2 University of Xxxxx Xxxxxxxx, Xxxxxx Xxxx, XX
3German Cancer Research Center (DKFZ), Heidelberg, Germany
4MD Xxxxxxxx Cancer Center, Houston, TX
5University of Minnesota, Minneapolis, MN
6Members of the Alliance through CTSU
7Pennsylvania State University/Hershey Medical Center, Hershey, PA
8Dana-Xxxxxx Harvard Cancer Center, Boston, MA
9Fred Xxxxxxxxxx Cancer Research Center
Contact information for Great Lakes-New England CVC:
2150 Cancer Center
University of Xxxxxxxx Xxxxxxx Xxxxxx
Xxx Xxxxx, XX 00000-0000
Telephone: (000) 000-0000 Fax: (000) 000-0000
Email: dbrenner @xxxxx.xxx (Pl) xxxxx@xxxxx.xxx (Lead CRA)
Contact information for the Data Management Coordinating Center (DMCC)
Xxxxxx Xxxxxxxx
EDRN DMCC Project Director
0000 Xxxxxxxx Xxx X, X0-X000
XX Xxx 00000
Xxxxxxx, XX 00000-0000
Phone: 000-000-0000 Fax: 000-000-0000 Email: xxxxxxxx@xxxxxxxxx.xxx
EDRN Biomarker Reference Laboratories (BRL)
University of Maryland-PI Xxxxxxx Xxxxx
Alliance for Clinical Trials in Oncology (Alliance)
Alliance- GLNE 010
Validation and Comparison of biomarkers for the Early Detection of Colorectal Adenocarcinoma
For any communications regarding this protocol,
please call the protocol resource person on the following page.
Study Chair:Xxx Xxxxxxx, M.D
Mayo Clinic
000 Xxxxx Xx. XX
Xxxxxxxxx, XX 00000
Alliance Protocol Resources
Questions: | Contact Name: |
Protocol document, consent form, | Xxxxxxxxxx X. Latky |
Regulatory issues | Research Base Research Protocol Specialist |
| Phone: (000) 000-0000 |
| Fax: (000) 000-0000 |
| E-mail: xxxxx.xxxxxxxxxx@xxxx.xxx |
* No waivers of eligibility per NCI
CLINICAL TRIALS SUPPORT UNIT (CTSU) ADDRESS AND CONTACT INFORMATION
To submit site registration documents: | For patient enrollments: | To Submit Study Data: |
CTSU Regulatory Xxxxxx 0000 Xxxxxx Xxxxxx, Xxxxx 0000 Xxxxxxxxxxxx, XX 00000 Phone - 0-000-000-XXXX Fax - 000-000-0000 |
Refer to Appendix H for specific instructions. |
All Groups must submit data via the EDRN’ s Validation Study Information Management System (VSIMS) . To obtain access for data entry, sites will be trained by webinar and given their own user name and password and access to the system.
For additional information about VSIMS, refer to the Manual of Operations, Appendix 13. For assistance with VSIMS or other data entry questions, call the VSIMS Helpline: 000-000-0000.
Do not submit study data or forms to CTSU Data Operations. Do not copy the CTSU on data submissions. |
Patient enrollments at all participating sites will use the GLNE’s Validation Study Information Management System (VSIMS). Refer to Appendix H for specific enrollment details.
Data management will be performed as follows:
All participating institutions will enter their data directly into the Early Detection Research Network (EDRN) supported VSIMS system of the Data Management and Coordinating Center (DMCC) as discussed in Section 4 of the protocol.
►Do not send study data or case report forms to the CTSU Data Operations. DO NOT copy the CTSU on data submissions.
Data query and delinquency reports will be sent directly to the enrolling site by the EDRN DMCC Operations Office. Please send query responses and delinquent data to the DMCC and do not copy the CTSU Data Operations.
CTSU sites should follow procedures outlined in Appendix H for Site Registration, Patient Enrollment, Adverse Event Reporting, and Data Submission.
For patient eligibility or treatment-related questions: Xxxxx Xxxx (000-000-0000 or xxxxx@xxxxx.xxx)
For questions unrelated to patient eligibility, treatment, or data submission contact the CTSU Help Desk by phone or e-mail:
CTSU General Information Line- 0-000-000-0000, or xxxxxxxxxxx@xxxxxx.xxx. All calls and correspondence will be triaged to the appropriate CTSU representative.
For detailed information on the regulatory and monitoring procedures for CTSU sites please review the CTSU Regulatory and Monitoring Procedures policy located on the CTSU members’ website xxxxx://xxx.xxxx.xxx
CTSU Web site is located at http s://xxx.xxxx.xxx
TABLE OF CONTENTS
1.0 | SUMMARY OF STUDY | 1 |
2.0 | SCHEMA USA Germany/Canada | 2 |
3.0 | OBJECTIVES | 3 |
4.0 | BACKGROUND AND SIGNIFICANCE | 3 |
4.1 | Current State of the Art: Recommended Early Detection | 3 |
4.2 | Current State of the Art: Serum Based Biomarkers for Colorectal Neoplasia | 4 |
4.3 | Rationale and Current State of the Art: Stool Based Biomarkers for Detection of Colorectal Neoplasia | 6 |
4.4 | Key Issues Driving Research Questions in CRC Early Detection Biomarkers | 8 |
4.5 | Rationale for Food Frequency Assessment and the Use of the NIH DHQ JI Food Frequency Questionnaire | 9 |
5.0 | STUDY DESIGN | 9 |
5.1 | Subject Recruitment | 9 |
5.2 | Eligibility | 9 |
5.3 | Study Procedures | 10 |
5.4 | Study Definitions | 11 |
5.5 | Biological Sample and Data Collection | 13 |
5.6 | Disclosure of results to subjects | 14 |
5.7 | Biomarker Analytical Approach | 14 |
5.8 | Data Collection, Management and Monitoring | 15 |
6.0 | STUDY CALENDAR (Table 1) | 15 |
7.0 | ANALYTICAL PROCEDURES | 16 |
7.1 | Vimentin methylation | 16 |
7.2 | Fecal immunochemical Test (FIT) | 16 |
7.3 | Galectin-3 Ligand | 16 |
7.4 | Circulating methylated genes BCATl/IKZFl (Clinical Genomics) | 16 |
7.5 | Hypomethylated LINEl from circulating cell free DNA (VolitionRx) | 16 |
8.0 | DATA ANALYSIS PLAN, SAMPLE SIZE JUSTIFICATION, AND STATISTICAL POWER. | 16 |
8.1 | Secondary Analyses | 18 |
9.0 | PROJECT MANAGEMENT PLAN | 19 |
9.1 | Strategies to Ensure Completion of Milestones | 19 |
9.2 | Timeline for Completing GLNE 010 | 19 |
9.3 | Endpoint Event Justification and Milestones | 20 |
9.4 | Endpoint Event Monitoring | 21 |
9.5 | Data Safety and Monitoring | 21 |
9.6 | Adverse Event Reporting | 22 |
10.0 | DATA MANAGEMENT | 23 |
10.1 | Registration | 23 |
10.2 | Timeliness | 23 |
10.3 | Completeness and Accuracy | 23 |
10.4 | Accuracy--Revisions and Corrections | 23 |
10.5 | On Site Data Audits | 23 |
10.6 | Sample Tracking | 24 |
10.7 | Confidentiality | 24 |
10.8 | Security | 24 |
11.0 | ETHICAL & REGULATORY CONSIDERATIONS | 24 |
11.1 | Institutional Review | 24 |
12.0 | REFERENCES | 25 |
13.0 | APPENDICES |
|
13.1 | Appendix A - Case Report Forms (separate document) |
|
13.2 | Appendix B - Recruitment Materials (separate document) |
|
13.3 | Appendix C - Blood SOP (separate document) |
|
13.4 | Appendix D - Stool SOP (separate document) |
|
13.5 | Appendix E- Colonoscopy SOP (separate document) |
|
13.6 | Appendix F -Model Informed Consent (separate document) |
|
13.7 | Appendix G- Urine SOP (separate document) |
|
13.8 | Appendix H -Alliance Site Logistics (separate document) |
|
Abbreviations and Definitions
Early Detection Research Network (EDRN)
Biomarker Reference Laboratory (BRL)
Standard operating procedures (SOPs)
University of Michigan (UM)
German Cancer Research Center (Deutsches Krebsforschungszentrum, DKFZ), Heidelberg, Germany
Deoxyribonucleic acid (DNA)
FIT test (sampling bottle provided by Polymedco for testing two different stool samples)
Data Management and Coordinating Center (DMCC)
Participant Identification Number (PIO)
Clinical Research Associate or study nurse (CRA)
Inflammatory Bowel Disease (IBD)
Hereditary non-polyposis colon cancer (HNPCC)
Familial Adenomatous Polyposis (FAP)
Validation Study Information Management System (VSIMS)
Fecal Occult Blood Test (Guaiac-based) FOBT
Immunoassay fecal occult blood test (FIT)
Department of Transportation (DOT)
Personal Health Information (PHI)
Data Safety Monitoring Committee (DSMC)
National Cancer Institute (NCI)
National Institute of Health Diet History Questionnaire II (DHQII)
Colorectal adenocarcinoma or adenomas with high grade dysplasia or adenomas greater than or equal to1 cm (Screen Relevant Neoplasia-{SRN})
Colorectal adenocarcinoma OR adenomas with high grade dysplasia (CRC/ HGD)
Cancer Trials Support Unit (CTSU)
Clinical Ligand Assay Satellite Services (CLASS)
1.0 SUMMARY OF STUDY
The goal of this trial is to estimate the sensitivity and specificity of stool vimentin methylation, serum galectin-3 ligand, and fecal immunochemical testing for 1) colorectal adenocarcinoma, or 2) screen relevant neoplasms (high-grade dysplasia or adenoma with 2:25% villous histologic features or adenoma measuring 2:1 cm in the greatest dimension or sessile serrated polyps measuring 1 cm or more in diameter) as single markers and in combination. Four thousand asymptomatic subjects aged 60 and older undergoing a first ever routine colonoscopic screening for colorectal cancer from U.S. community and major medical center outpatient settings across multiple centers and consortia will be recruited. An additional five thousand subjects age 50 and older undergoing routine colonoscopic screening for colorectal cancer will be recruited in Germany and Canada (non-US sites). Up to 9,000 subjects will be recruited in this protocol, adding to the 4,677 confirmed and evaluable subjects already recruited. Subjects will meet with research staff prior to initiation of any colonoscopic preparative procedure. After completing informed consent, they will complete Early Detection Research Network (EDR.i’-J) data element forms. Blood and urine will be obtained following EDR.i”‘J standard operating procedures (SOPs). Subjects will be provided with kits to collect stool samples for fecal immunochemical test (FIT) and stool tests. The collected samples will be shipped to the Central Laboratory at the University of Michigan or German Cancer Research Xxxxxx (Xxxxxxxxx Xxxxxxxxxxxxxxxxxxxxxx , XXXX), Xxxxxxxxxx, Xxxxxxx where the stool will be homogenized , aliquoted, and stored at the Umiversity of Michigan CLASS laboratories. The FIT tests will be sent to the Central Laboratory at the University of Michigan or to DKFZ for quantitative analysis following standard operating procedures provided by Eiken Chemical Company. Data from the screening colonoscopy will be obtained. One year after colonoscopy, subjects will be contacted to determine if they have had a neoplastic colorectal diagnosis or other neoplastic events. Data management and protocol coordination will be performed by the Data Management and Coordinating Center (DMCC) of the EDRN along with the GLNE Prevention Research Base at the University of Michigan and will include a Web-based front end and relational database backend, with biosample tracking (VSIMS). Biosamples will be managed in a high quality repository facility at the University of Michigan until shipment to the EDR.”I\T repository at NCI at Xxxxxxxxx Central Repository and to analytic partners.
We will estimate sensitivities and specificities and the corresponding confidence intervals of the stool DNA tests and serum/plasma tests for detection of invasive colorectal neoplasms and for screen relevant neoplasias (Aim 1). We will then test the primary hypothesis to confirm the clinical accuracy of a particular biomarker test or panel (Aim 2). The specific primary hypothesis will be defined prior to data analysis based on state of the art information available at that time about candidate biomarkers and tests. Several specific examples of potential primary hypotheses are given to justify study sample size. Finally, several alternative tests and multi-marker panels will be evaluated. (Aim 3). In secondary analysis, we will (a) provide measures of diagnostic accuracy standardized to the age and gender distribution of US population and (b) assess the effect of subject heterogeneity on the marker performance. A primary objective is to establish an archive of appropriately preserved stool, serum, plasma and DNA human biospecimens to be used by ED -approved investigators for future validation and biomarker discovery research (Aim 4).
1
Adults, age 60 and older who are undergoing their first ever screening co1onoscopy and are wi11ing to participate |
| Adults, age 50 and older who are undergoing their first ever screening colonoscopy and are willing to participate | |||||||||
|
|
|
|
|
|
|
| ||||
Eligible
Willing to sign Informed Consent document Able to tolerate removal of 50 ml of blood (5 tubes or 3.5 tablespoons) Willing to collect 2 stool samples Never had a full colonoscopy for screening purposes
|
Ineligible
Inability to provide informed consent History of Inflammatory Bowel Disease Overt rectal bleeding within 1 month Positive FOBT or FIT in the past 12 months Undergone resection of the colon for any indication Subjects with known HIV or chronic viral hepatitis (Hepatitis B and C) Subjects with known or suspected HNPCC (Xxxxx Syndrome) or FAP Any cancer within 5 years prior to enrollment except squamous cell carcinoma of the skin or Basal cell carcinoma of the skin Prior history of Colon Cancer or Rectal Cancer.
| ||||||||||
|
| ||||||||||
| BASELINE OR PRE-COLONOSCOPY
Signed Informed Consent Blood Collection (50 ml) Urine Collection(100 ml) Complete Questionnaires Stool collection and FIT (x2) and shipped to UM/DKFZ Compensation sent to subject
|
| |||||||||
|
|
|
| ||||||||
| COMPLETE COLONOSCOPY
Endoscopy and pathology reports collected If eligible, compensation for enrollment sent to site/or credits earned Central pathology review of invasive colorectal neoplasms Central pathology review of screen relevant neoplasias
|
| |||||||||
|
|
|
| ||||||||
|
Follow up phone call one year after colonoscopy & provide any additional Surgery/Pathology data
|
| |||||||||
|
|
|
| ||||||||
| Review of relevant medical records from follow-up events
|
| |||||||||
2
We propose a prospective cross-sectional PRoBE-compliant validation trial of stool-based and serum-based tests for the detection of colorectal neoplasia (3). The trial is powered to evaluate tests for detecting early stage colorectal adenocarcinoma. This is the most stringent, conservative approach to the early diagnosis of colonic neoplasia and addresses the most important endpoint of identifying individuals with curable, early stage cancer.
Aim 1: To estimate the sensitivity and specificity for 1) colorectal adenocarcinoma or 2) screen relevant neoplasms (high-grade dysplasia or adenoma with 2’.:25% villous histologic features or adenoma measuring 2’.:1 cm in the greatest dimension or sessile serrated polyps measuring 1 cm or more in diameter) of the following individual colorectal neoplasia early detection biomarkers using colonoscopy as the gold standard:
stool vimentin methylation
serum galectin-3 ligand
fecal immunochemical tests (FIT)
Circulating methylated genes BCATl /IKZF l (Clinical Genomics)
Hypomethylated LINEl from circulating cell free DNA (VolitionRx)
Other currently unspecified biomarkers
Aim 2 (primary objective): To assess the accuracy and potential clinical value of a test for detection of colorectal adenocarcinoma. The specific test and relevant hypothesis are not defined now but will be chosen when all samples have been collected. This will allow the primary hypothesis to incorporate all information about markers and clinical practice that is available at the time of analysis and will ensure that the most compelling and timely hypothesis is tested Statistical power calculations demonstrate that the study is well powered for these hypotheses (Section 8.0).
(Secondary objective): While the study will be powered for the primary objective, we shall also carry out a similar assessment of potential utility of a clinical test for SRN.
Aim 3
To validate, or to construct, a combined early detection biomarker panel using the above individual biomarkers (stool vimentin methylation, serum galectin-3 ligand, FIT, circulating methylated genes BCAT l /lKZFl , hypomethylated LINEl from circulating cell free DNA), and other unspecified future biomarkers, and describe its performance for 1) colorectal adenocarcinoma and for 2) screen relevant neoplasms.
Aim 4
To establish an archive of appropriately preserved stool, serum, plasma and DNA human biospecimens to be used by EDR: -approved investigators for future validation and biomarker discovery research.
4.0BACKGROUND AND SIGNIFICANCE
4.1Current State of the Art: Recommended Early Detection
Randomized controlled trials have shown that annual or biennial fecal occult blood tests (FOBT) reduce colorectal cancer (CRC) mortality by 15% to 33% (4-6). The reduction is durable over 3 decades (7). Population based cohort studies of colonoscopic screening demonstrate reduced CRC mortality, primarily in distal but not in the proximal colon (8-10). This discrepancy has been attributed to endoscopic quality issues, the technical difficulties in detecting lesions in the right colon, and the more frequent occurrence of flat and depressed dysplastic lesions in the right colon (11-14). In tandem colonoscopy studies, a subset oflarge polyps may be missed by a single examiner. Shorter withdrawal is time linked to a lower adenoma detection rate (15, 16).
Flat and depressed lesions are more challenging to detect and have been described with a relatively high prevalence in a US colonoscopy cohort (17). While colonoscopic removal of adenomatous polyps reduces CRC mortality (18), prospective, randomized controlled trials of screening colonoscopy have been initiated by the VA and in Europe (18-20). Over-diagnosis (i.e. early detection of indolent invasive neoplasms that do not cause mortality) or lead-time bias in early detection of colorectal neoplasms do not degrade the efficacy of screening and early detection for colorectal cancers (21).
3
Current screening guidelines for average risk individuals vary world-wide. In the United States the American Gastroenterology Association recommends testing for early detection of adenomas and cancer (structural examination) or of cancer (non-invasive stool tests) beginning at age 50 (22). The United States Preventive Services Task Force (USPSTF) recommends fecal occult blood testing (FOBT) every two years with optional endoscopic screening with either flexible sigmoidoscopy or colonoscopy (23). The majority of developed countries recommend fecal occult blood testing every two years but do not support endoscopic screening (24); albeit with some exceptions (e.g. Germany (24, 25)). In 2012, 65.1% of the United States adults adhered to USPSTF colorectal screening guidelines with colonoscopy the commonly used screening method (61.7%) followed by FOBT (10.4%) (26) whereas colonoscopic screening adherence in Germany is 16% (25). Over 20 years of SEER data (1991 to 2011), United States CRC incidence (all races, males, females) has fallen from 59.5 cases in 1991 to 39.3 cases per 100,000 in 2011 (35% reduction) with a corresponding mortality reduction over the same time period from 24.0 to 15.1 deaths per 100,000 (37% reduction) (27). Widespread adherence to screening guidelines in the United States may be driven by the profound changes in the organization of medical care including enhanced access via the Affordable Care Act, rigid guideline enforcement by payers with physician performance incentives and disincentives, and the rapid adaptation of electronic medical record systems enabling ease referrals for screening, compliance reminders, and management tracking of compliance to care guidelines (28).
4.2Current State of the Art: Serum Based Biomarkers for Colorectal Neoplasia
Reasons for non-adherence with stool based or colonoscopic based CRC screening include the volume of bowel preparation, inadequate analgesia, no recommendation from primary physician, embarrassment (29) or cultural taboos surrounding collection or manipulation of stool provide rationale for discovery and validation of circulating biomarkers for early detection of colorectal neoplasia. Circulating signatures may be detected from neoplasm generated genetic products, antigens, antibodies, glycans, circulating tumor cells.
4.2.1Genetic Products
In a recent study of 24 CRC patients, mutant DNA fragments (circulating tumor DNA, ctDNA) are found in at relatively high concentrations in the circulation of most patients with metastatic cancer and at were detected in ~70% of patients with localized cancers (30). The direct detection of aberrant genes or genetic material specific to colorectal neoplasms (e.g. APC, -catenin, K-ras, DCC, and p53) has been limited by the technical challenge of DNA recovery, the large number of potential underlying genetic mutations, and by the limited sensitivity of any single genetic alteration due to the extremely low abundance gene mutations in circulating plasma or serum (30-35). DNA hypermethylation, in contrast, affects residues in regulatory portions of genes and provides major advantages in designing biomarker assays (34, 36-38). Digital based quantitative technologies improving upon bisulfite conversion while minimizing bisulfite associated DNA fragmentation and single molecule detection technologies (39) permit cost effective development of DNA hypermethylated gene biomarkers. Such technology detected circulating methylated vimentin with 59% sensitivity (39). Septin9, a methylated gene discovered in tissues with array technology (40, 41), detects CRC with 50% sensitivity and 92% specificity in a large (7941 participants) prospective colonoscopy verified screening trial (1). For early stage CRC, Septin9 sensitivity decreased to 35%. While circulating methylated CpG DNA promoter sites appear to have higher CRC detection performance than other genetic detection strategies, they substantially lag behind stool based detection of blood DNA markers or endoscopy. Nevertheless, for individuals refusing to use stool based screening, detection sensitivity of circulating methylated DNA markers appears equivalent to guaiac based stool screening and has the potential advantage of capturing the 35% of the population refusing stool screening. miRNAs are stable and detectable in serum and plasma. As in stool, numerous up and down regulated xxXXX stool signatures discovered using unsupervised array technology may be useful as CRC detection biomarkers. A recent review identifies 19 miR..”l’,,XXx as individual or groups in panels as candidates for detection markers; but, insufficient clinical validation renders the data generated to date using small convenience sets confusing and not mechanism driven (42).
4.2.2Proteins
Antigens: Approximately 50% of all proteins are estimated to be glycosylated (43). Glycan abundance and their micro- and macro-heterogeneity can be changed in a disease-specific manner (44). Glycoprotein screening studies, many EDR.”N s upported , have relied on immunoprecipitation or lectin affinity capture of whole glycoproteins and mass spectrometry identification of the de-glycosylated protein portion or probed with lectins in an array format containing up to a few hundred antibodies (45-49). Sialylated Xxxxx A and Xxxxx X moieties carrying proteins identify panels of potential markers. The Xxxxx EDRN laboratory has found seven such proteins (B3GNT5, CD44, HSPG2, IL6, INHBC, NOTCH4 and VWF) which, when combined in discovery set plasma samples ROC AUC of 0.83 (50). GLNE discovered glycan ligand, galectin-3 ligand is circulating glycan biomarker in large population based prospective validation (51).
4
Antibodies: Serum antibodies recognizing multiple colon cancer antigens can be detected in colorectal adenocarcinoma patients’ markers (52-54). Preliminary validation of single or small autoantibody panels have been disappointing (55). For example, antibodies to the Fas receptor have 17% sensitivity when 100% specific for CRC detection (56). Experience with p53, Hsp60, and nucleobindin 1 (Calnuc) autoantibodies has been better (~50% sensitivity/70 to 90% specific); but, they are not specific to CRC (55, 57, 58) and cannot be used as a colon specific screening tool. Discovery sets that include a miniarray of autoantibodies with other markers have reported improved detection accuracy (sensitivity 83%/specificity 90%) (59) but require clinical validation.
Cytokines/growthfactors: High serum concentrations of insulin-like growth factors (IGF) and low levels of their binding proteins have been shown to correlate with CRC risk in large cohort studies (60-63) but have low sensitivities with high specificities for CRC detection. Other cytokines or angiogenesis factors such as XXX-X 0 (00-00), XXXX (00, 00), xxxxxxxxxx (73), endostatin (74), and endothelins (75, 76) also have low sensitivity in small convenience sets and have not proceeded to clinical validation.
Other proteins: Of the matrix metalloproteinases (77-79), plasma TIMPl is elevated in CRC but has not had sufficient sensitivity in larger validation trials to merit development as a detection biomarker (80). Cell adhesion molecules (81) have low sensitivities for detection of early stage CRC.
4.2.3Circulating Tumor Cells
Circulating tumor cells (CTCs) entering the vascular space from primary neoplasms have been considered to be initiators of metastases (82-84) and can be detected in early stage invasive neoplasms (85, 86). CTC isolation from epithelial cancers initially used antibody capture technology dependent upon epithelial adhesion (EpCAM) and cytokeratins (82).
This technology limits CTC detection of early stage neoplasms because CTCs are thought to undergo epithelial to mesenchymal transition (EMT), epithelial traits are lost and epithelial marker such as EpCAM and cytokines are downregulated. CTCs present in as few as 1 cell in 5 x 109 red cells, and up to 5-10 x 106 white blood cells, are rare events (84). Newer microfluidic or centrifugation devices appear to more efficiently capture CTCs (85, 87). The inclusion of mesenchymal/EMT-specific antibodies, for example, vimentin, PLS3 may improve CTC capture and/or expansion (84). With the emergence of ex-vivo expansion protocols of CTCs and the increased ability to detect stem like or stem progenitor cells, CTCs are of future interest as an early cancer detection diagnostic (85, 87), but remain in the technology development phase.
4.2.4Special consideration-EDRN discovered and preliminarily validated circulating biomarker: Galectin-3 Ligand ELISA as a Serum Biomarker for the Detection of Colorectal Neoplasia
The galectins are widely distributed and evolutionarily conserved carbohydrate binding proteins characterized by their binding affinity for p-galactosides and by conserved sequence elements in the carbohydrate-binding region (88). Galectin-3 is the galectin that is of most interest in regard to colon cancer because of its demonstrated role and cancer progression and metastases and interaction with mucins(89-93). Galectin-3 ligands include laminin, LAMP-1 and 2, LPS and colon cancer mucin. The major galectin-3 ligand detected in serum is a 40 kDa band distinct from MUC2 and other mucins CEA, and Mac- 2-BP. We reported a true positive rate for the detection ofCRC of 91% and false positive rate of 18% using preliminary data using quantitative Western blot technology on a convenience set of GLNE serum (51).
We developed a sensitive, reproducible ELISA assay for galectin-3 using a new antibody we created. This was used to assay the GLNE colorectal reference set (50 colorectal adenocarcinomas/50 adenomas/50 endoscopically normal controls). The ROC analyses for galectin-3 ligand combined with FOBT (fecal occult blood test-guaiac based) for detection of colorectal adenocarcinoma versus controls who had normal colonoscopy shows an area under the ROC curve of 0.91, while galectin-3 ligand detection of colorectal adenocarcinoma alone versus controls who had normal colonoscopy shows an area under the curve of 0.84. The true positive rate of galectin-3 ligand with FOBT for detection of CRC is 64% with a false positive rate of 5%. Without FOBT, true positive rate of galectin- 2 ligand was 72% with a false positive rate of 20%.
5
4.3Rationale and Current State of the Art: Stool Based Biomarkers for Detection of Colorectal Neoplasia
4.3.1Occult blood tests
Stool testing as a screening approach offers the potential advantages of noninvasiveness, low cost, avoidance of cathartic preparation, and minimal impact on work time or daily activities. guaiac based FOBT is not specific for human blood, and consequently it has a high false positive rate for colorectal neoplasia. The fecal immunochemical test (FIT) detects human hemoglobin, thus eliminating the false positives caused by non human hemoglobin in the diet (94, 95). FIT tests are more sensitive at detecting CRCs (sensitivity range 61% to 91%) and adenomas (sensitivity range 16% to 31%) than classical unrehydrated guaiac FOBT (Hemoccult II) (sensitivity range 25% to 38% for CRC; 16% to 31% for advanced adenomas) (96, 97). A recent meta-analysis that analyzed data from 19 prospective randomized trials or cohorts using 8 different commercially available FIT tests with colonoscopy or 2 year observation endpoints reported an overall sensitivity for detection of CRC of79% (95% CI = 0.69-0.86), specificity of 94% (95% CI = 0.92- 0.95) and overall accuracy (defined as hierarchical summary receiver operating characteristic (ROC) curve) of 95% (95% CI= 93% - 97%) (Figure 1). Differences in performance characteristics among FIT brands were small, particularly between the two major brands used OC-Light (Eiken Chemical) and QC-Micro/Sensor (Polymedco + Eiken Chemical). The Polymedco product is widely used in the USA. Quantitative FIT (Eiken OC-SENSOR) >177 µg/gm stool combined with age and sex predicts 11.46 fold risk of a large adenoma over lower risk groups (98).
| Fig 1 from Xxx et al (2): Hierarchical ROC curve of the sensitivity versus specificity of FIT. The diamond= summary point of the curve to which the pooled sensitivity and specificity correspond. Dashed line = 95% Cl for summary point; dotted line = 95% confidence area of FIT diagnostic accuracy. AUC = area under the curve; SENS = sensitivity; SPEC = specificity. |
4.3.2Stool DNA tests
Since the neoplastic transformation process of the colonic epithelium results in cells shedding into the stool, collection of fecal material is likely to yield detectable molecular and biochemical events associated with cellular transformation (99, 100). First generation multi-marker stool DNA tests detected 52-73% of CRCs, 41-49% of CRCs plus adenomas with high grade dysplasia, and 15-46% of adenomas 2:1 cm, with specificities of 84-95% (101, 102). Stool DNA test performance in both studies was compromised by failure to use stabilization buffer with stool collection, inefficient marker recovery from stool, and relatively insensitive analytical methods. Exact Sciences modified their previously published stool DNA panel (102) and now uses a panel consisting of methylated BMP3 and NDRG4 promoter regions, mutant K-ras (7 point mutations, Exon 2, codons 12,13), and a proprietary FIT test). In a recently published cross sectional validation study of 9,989 patientsundergoing screening colonoscopy, the panel performed with a sensitivity of 92% for CRC; 84% for CRC + high grade dysplasia; and 42% for advanced adenomas (Figure 2) (103).
The specificity was 87% for CRC, the ROC AUC for the Exact Sciences DNA stool panel for the detection of colorectal cancer is 0.94. FIT alone (Polymedco FIT) performed with sensitivity of 73.8% and specificity of 94.9% for detection of CRC and sensitivity of 23.8% for screen relevant neoplasia. Stool DNA component of the panels adds ~20% sensitivity to FIT. The USPSTF is currently assessing the role and contribution of fecal DNA panels such as the Exact Sciences panel to CRC screening (104).
6
| Fig 2 from lmperiale et al [lmperiale 2014 #5977) sensitivity for detection of CRC by Exact Sciences stool DNA panel + FIT (ight blue) vs Fit alone (dark |
4.3.3Vimentin Methylation as a Stool DNA Test
Aberrant methylation of vimentin exon 1 was initially described as a highly frequent biomarker of colorectal cancers and adenomas by Xxxxxxxxx and co-workers (105). In reproducible studies, aberrant methylation of vimentin has been detected in 72%-83% of colon cancers and 70%-84% of colon adenomas (105, 106). The current assay for detection of vimentin exon 1 methylation is based on using methylation specific PCR (MSP). Adaptation of the vimentin MSP to testing fecal DNA is accomplished by recovery ofvimentin DNA sequences from human stool using hybrid capture to vimentin specific oligonuclotides(l05). Initial study showed that MSP assay of vimentin purified from feces (fecal vimentin DNA) detected methylated fecal vimentin DNA in 46% of cancer patients (N=94) at a specificity of 90% (N=198)(105). This initial study involved collaboration between the Xxxxxxxxx laboratory who had discover d the methylated vimentin DNA marker, and Exact Sciences, who implemented detection of this marker in fecal DNA. This initial study was limited by use of samples that had suffered problems of DNA degradation during sample collection and shipping (102). A recently published two stage followup study lead by Xxxxxxxxx et al in collaboration with Exact Sciences and the Xxxxxxxxx laboratory showed markedly improved results with the use of a DNA stabilizing buffer added to stools at the time of collection (107). Detection of methylated fecal virnentin
DNA was found in 77% of cancers (N=82) at 83% specificity (N=363). Six of 7 adenomas with high-grade dysplasia were also detected. This assay has successfully detected 55% (N=22) of adenomas that were greater or equal to 1cm in size (107). This is a published assay of capture of fecal vimentin DNA and then MSP detection of methylated vimentin exon 1 sequences (105, 107, 108).
4.3.4Other Stool Based Biomarkers Under Investigation
Considerable interest in fecal microbiome populations has triggered EDRN supported investigators into identifying unique bacterial species that are associated with colonic carcinogenesis and suggests that a microbiome signature may be a useful stool biomarker for CRC risk (109, 110). Metabolome signatures promise to identify amino acid or fatty acid profiles associated with colorectal cancer or high risk (111) have been preliminarily developed in EDRN supported research. Micro-RNAs (miR._NA) have both oncogenic and suppressor properties, can be detected in stool, and have been explored as stool based early detection biomarkers (112, 113). Studies published to date have used small convenience samples and array technologies that have identified diverse and non-reproducible miRNAs as classifiers for colonic neoplasms.
7
4.3.5Urine base Biomarkers
We demonstrated previously that human urine contains circulation-derived DNA[< 300 base pairs (bp), designated as low molecular weight (LM\V) DNA] and that LM\V urine DNA can be used to detect colorectal cancer (CRC) associated k-ras mutations from patients with CRC. A quantitative MethylLight PCR-based assay targeting a 39-bp template of the hypermethylated vimentin gene (mVIM) was developed to detect circulation derived mVIM DNA. A blinded concordance study was performed using matching tissue and urine DNA samples from patients with CRC. The 20 CRC tissue samples and 20 urine samples from patients with CRC were provided with barcodes. LMW urine DNA and tissue DNA were isolated, bisulfite converted and assayed for mVIM. The mVIMwas detectable in 85% (17/20) of the CRC tissue DNA samples and 75% ofLMW urine DNA. As control, LMW urine DNA isolated from 20 subjects with no known neoplasm was also tested for the mVIM DNA. Two of 20 (10%) normal control LMW urine DNA contained detectable mVIM DNA. After all of the samples were tested, the urine and tissue ID numbers were unblinded and matched The concordance value between the mV/J\1-positive CRC tissue and matched urine DNA samples was 71% (12/17). We thus conclude that CRC-associated mVIM DNA can be detected in the urine of patients with CRC with a concordance of 71% between marker-positive tissue and matched urine samples with a sensitivity of 75% (Su Y-H, personal communication). These results support further development of a urine test for CRC screening.
4.4Key Issues Driving Research Questions in CRC Early Detection Biornarkers
Until therapeutic agents with much greater potency and minimal side effects are developed, the current best strategy for reducing cancer morbidity and mortality is early detection of neoplastic disease (114). Key opportunities in the current state of colorectal screening and early detection include:
1.Enhancing adherence to current screening guidelines: Screening and early detection reduce mortality from colorectal cancer; yet 35% of the population in the USA remain non-adherent. Adherence is much lower in other countries (25). The barriers to these recommendations (cost, discomfort, cultural taboos) may be overcome with circulating biomarkers that provide individuals with persuasive evidence that undergoing invasive screening procedures, i.e. colonoscopy, will have important life-saving benefit that reduces mortality from CRC (8-10, 18). Developing, validating and bringing circulating biomarkers to population screening use remains a high priority that will likely increase adherence to endoscopic screening. GLNE 010 addresses this priority by working closely with EDRN and industry groups to clinically assess and validate circulating biomarkers of CRC risk that might drive individuals who might decline to endoscopic screening.
2.Tailoring colonoscopic screening to individual risk: Recently published data from the Clinical Outcomes Research Initiative found the prevalence of large polyps higher in blacks than whites among both men and women (115). Tailoring endoscopic screening to those at risk while limiting screening for those with minimal or no risk (116, 117) will enhance screening adherence and eliminate excess cost. Recommendations for tailoring were primarily population demographic based (116, 117); yet, the translation of carcinogenesis biology and genetics into biomarker panels with extremely high sensitivity (99%), i.e. no false negative tests, promises precise tailored endoscopic screening. The current state of art stool using based biomarker tools is coming close-92% sensitivity (103) but insufficient to permit tailored or individualized risk. GLNE 010 addresses the priority ofbiomarker driven tailored risk by completing the ongoing phase 3 validation trial of stool and circulating biomarkers and using the extensive repository created by this and other GLNE protocols to rapidly identify new markers that may enhance sensitivity of the current biomarker panels.
3.Persistently positive stool DNA tests with negative colonoscopic screening: The stool methylated DNA panels report 5% false positives (103, 108). A positive stool DNA test with a negative screening colonoscopy could potentially arise from neoplasia in the upper gastrointestinal tract or from occult and missed lesions in the colorectum. The latter is a particular concern in the right colon, where flat lesions and/or sessile serrated adenomas are more prevalent. Preliminary data from the Case Western EDRN BDL found near 100% vimentin methylation in gastric dysplasia while no methylation in adjacent gastric mucosa (X. Xxxxxxxxx, Personal Communication). In Xxxxxxx’x esophagus (BE), 7 of 7 high grade dysplasias (HOD), and 15 of 18 esophageal adenocarcinomas (EAC) and even in some squamous cancers (SCC) had methylated vimentin , whereas it was absent in all 9 normal squamous mucosa (118). A “false positive” stool DNA test may detect dysplasia or invasive neoplasms in the upper GI tract. The GLNE will propose to address this priority in the future in a future project. This project, to be submitted as a separate proposal will propose a longitudinal study of participants registered in an ongoing cross sectional Phase 2 colon biomarker validation trial with a positive stool test and negative colonoscopy registered in the current
8
4.5Rationale for Food Frequency Assessment and the Use of the NIH DHQ II Food Frequency Questionnaire
Numerous epidemiologic studies have controversially implicated high total fat and saturated fat, alcohol, inadequate calcium, vitamin D, dietary fiber, vitamin B6, folate, methionine, antioxidant vitamins such as C, and E, and lack of fruits and vegetables as dietary risk factors for colon carcinogenesis (119-127). The causal contribution of these dietary factors to risk of colon cancer has been difficult to assess and compare in meta analyses due to different instruments used to assess diet, including diet records, 24 hr. dietary recalls and food frequency questionnaires (128, 129). However, because there is significant epidemiologic support for dietary variables affecting cancer risk in populations (130-132), it is important to collect dietary information along with human biosamples to allow future study of the relationship of selected dietary variables and their impact upon biomarkers of cancer risk, for early detection, or post diagnosis prognosis. For example, vimentin methylation is a key stool DNA marker we propose to validate as an early detection tool in this trial. Since the methylation reaction requires methyl tetrahydrofolate (133-135), it is conceivable that dietary folate may impact the methylation status of this and other future methylated biomarkers for cancer risk and detection.
In a large validation biosample and annotated data set such as the one in this trial, diet intake among different subjects is likely to be an important source of bias, thus an adjustable variable in the analysis of validated biomarkers. We have chosen to administer a food frequency questionnaire to the subjects emolled in this trial because it assesses dietary exposures over time (typically 6 months). We recognize the weaknesses of a food frequency instrument, including recall bias, but the instrument has value when used as a semi-quantitative measure to rank order individuals according to their intake of a given nutrient rather than a continuous variable (136, 137). We propose to use the computerized self-administered user-friendly National Institute of Health Diet History Questionnaire II (DHQII). The DHQ I was developed specifically to study dietary risk factors for cancer and has been validated to adequately assess dietary intake over time against established FFQs such as the Block and Xxxxxxx FFQ (136, 138). The questionnaire assesses supplement use in addition to dietary intake data and has been used in multiple cancer studies to date. The DHQ II is a refinement of the validated DHQ I with improved separation of some food sub categories to enhance detail of data intake.
5.0STUDY DESIGN
5.1Subject Recruitment
The clinical research associate or study nurse (hereafter “CRA”) at each clinical site will identify subjects with appointments for colonoscopy via !RB-approved HIPAA-compliant site-specific methods (Appendix B-tailored to each site). Recruitment methods could include letters from the primary care physicians and gastroenterologists, direct referrals to the study team by physicians, in-clinic recruitment advertisements, use of navigator programs, county or statewide screening programs, and other !RB-approved means of identifying and contacting subjects. Interested subjects will be asked to participate in a baseline visit prior to initiation of colonoscopy preparative procedures, either at the local Center or during a visit to the subject’s home by a CRA. Advertisements (e.g., newspapers, AARP Magazine, Xxxxxxxxxxxxxx.xxx) may also be used to recruit subjects from the surrounding communities.
5.2.1Inclusion Criteria (at time of consent)
Subjects at US Sites
Adults 60 and older
Never had a full colonoscopy for screening purposes
Willing to sign informed consent
Able to physically tolerate removal of about 50 ml of blood
Willing to collect 2 stool samples
Subjects at Sites in Germany/Canada
Adults 50 and older
Never had a full colonoscopy for screening purposes
Willing to sign informed consent
Able to physically tolerate removal of about 50 ml of blood
Willing to collect 2 stool samples
9
5.2.2Exclusion Criteria (at time of consent)-All subjects
Inability to provide informed consent
History of Inflammatory Bowel Disease
Overt rectal bleeding within 1 month (30 days) (including due to suspected hemorrhoids)
Positive guaiac-based occult blood or fecal immunochemical test (e.g. FOBT, FIT) in the past 12 months (365 days)
Undergone resection of the colon for any indication
Subjects with known HIV or chronic viral hepatitis (Hepatitis Band C)
Subjects with known or suspected HNPCC (Xxxxx Syndrome) or FAP
Any cancer within 5 years prior to enrollment except squamous cell carcinoma of the skin or Basal cell carcinoma of the skin.
Prior history of Colon Cancer or Rectal Cancer.
5.3.1Enrollment and Registration Procedure
Subjects who meet the eligibility criteria will be scheduled for a baseline visit. The baseline visit must occur prior to any preparative regimen for colonoscopy (e.g. PEG {Golytely, Halflytely}, Miralax/Gatorade, Suprep, etc.) and within 16 weeks of the scheduled colonoscopy procedure. At this baseline visit, subjects will provide informed consent (see model consent, appendix F) for analysis of stool, urine and blood samples for biomarkers; medical record review, including colonoscopy and pathology reports; and for completion of questionnaires.
The subject will be enrolled and given a unique participant identification number (PID) generated randomly by the DMCC. The sites will subsequently link the PID to the specimen collection kits once specimens are collected.
5.3.2Demographic and Other Data Collection
The subject will be asked to provide data to complete EDRN demographic and medical history questionnaires. Clarification or additional information may be obtained from the medical records. Case report forms (CRFs) will also be used to collect information on concomitant medications, colonoscopy outcomes, resection information, any new cancer treatment, and new diagnostic tests. Long term data collection (medical records review and follow up data) will be prompted by information gathered at a phone call with the subject at one-year post colonoscopy. Data may be collected via face-to-face interviews, via phone or email interviews, or returned by mail dependent on subject preference. Subjects (U.S and Canada only) will be asked to complete a NCI DHQ II food frequency questionnaire (Appendix A) at home after the baseline visit as defined in Section 5.3.1. The NCI DHQII can be done online through a secure web-based system ( xxxx://xxxxxxxxxx.xxxxxx.xxx/xxx0 ). If subjects chose to report diet data on paper forms, sites will be responsible for entering the data into the web-based system.
5.3.3Sample Collection: Blood
Baseline blood samples will be obtained according to standard operating procedures (Appendix C). The blood will be collected during or after the baseline visit but prior to any preparative regimen or procedure as detailed in section 5.3.1 and the Operations Manual. Samples must be collected within 16 weeks prior to the qualifying colonoscopy (detailed in the operations manual). Blood may not be collected at or after the colonoscopy.
5.3.4Sample Collection: Urine Sample
A baseline urine sample will be obtained according to standard operating procedures (Appendix G). The urine will be collected during or after the baseline visit, but prior to any preparative regimen or procedure as detailed in section 5.3.1 and the Operations Manual. The urine specimen must be collected within 16 weeks prior to the qualifying colonoscopy. Urine may not be collected at or after colonoscopy.
10
5.3.5Sample Collection: Stool Sample and FIT #1
Subjects will be required to collect stool samples prior to any preparative regimen or procedure within 16 weeks prior to colonoscopy. Women will be asked to avoid collection during heavy menses if applicable. Subjects will be asked to collect their stool in the collection bucket (hat) provided. Subjects will be given detailed instructions and complete kits to collect the stool samples at home. They will prepare an OC_SENSOR FIT (Eiken Chemical Company) (FIT #1) from the stool sample. Subjects will also collect scoops of stool into a container with an EDTA-based buffer (“buffered stool”) and additional scoops of stool into tubes provided to be sent on ice packs (“native stool”) The subjects will then package both the stool and the FIT for shipping per provided instructions. The US and Canadian subjects will ship the stool sample to the Central Laboratory at the University of Michigan using pre-paid DOT (Department ofTransportation)-compliant packaging.
German subjects will send their stool samples to the German Cancer Center (DKFZ) (Xx. X. Xxxxxxx).
5.3.6Sample Collection: FIT #2
Subjects will be asked to collect another bowel movement (ideally the next one) for a second FIT only (FIT #2). The subject will use the 2nd FIT to collect another sample from the stool collected on paper provided. The subject will mail the FIT using provided self addressed postage-paid envelopes. The US and Canadian subjects will ship the FIT #2sample to the Central Laboratory at the University of Michigan and the German subjects will send their stool samples to the German Cancer Center (DKFZ) (Xx. X. Xxxxxxx).
5.3.7Subject Compensation
To compensate for the inconvenience and cost of driving and parking, $25 will be provided to each subject once blood samples, urine samples, stool samples and questionnaires are completed. If the research coordinator visits the subject at home, no payment will be offered at the site’s discretion. U.S. and Canadian recruiting sites will receive gift cards to distribute to subjects that complete the requirements to receive payment. Gift cards will be to places like Target, Walmart, or other similar stores in the specific region, purchased by UM Prevention Research Base staff and distributed to sites. Sites are required to account for distribution of gift cards to subjects. German subjects will be paid according to local policies.
5.3.8Colonoscopy Standards
Colonoscopy standards for inclusion of the data into GLNE 010 will include verification of insertion to cecum, photos of all lesions (available at the site), size, histology and location in colon of all suspected colorectal cancers, adenomas, or other polyps. Case report forms will capture some of this information; the rest will be reviewed directly at site monitoring visits or by review of redacted reports.
5.3.9Sample Collection: Tissue Samples
One H & E slide from clinical tissue blocks of all detected colorectal adenocarcinomas , high grade dysplasia and advanced adenomas will be obtained (given, shared digitally or borrowed), sent to the lead site and reviewed by a reference GI pathologist at the University of Michigan. Up to ten slides (lOum thick sections) from clinical tissue blocks of all detected colorectal adenocarcinomas, high grade dysplasia and advanced adenomas will be obtained whenever possible and sent to the lead site for storage. Specific details will be worked out with each site depending on costs and standard practices.
5.3.10Sample Labeling and Tracking
All samples will be labeled with a unique specimen ID (embedded barcodes or other labels) managed by the DMCC. The site will subsequently associate the specimen IDs to the PID. The bar codes will be scanned at each step of the procedure (collection, on-site processing, shipment, receipt, and storage in a repository). All biosamples are property of the EDRN.
11
5.4.1Assessing Inclusion Criteria-Definition of “Full Colonoscopy”
Prior colonoscopy eligibility requirements for screening (versus surveillance) indications are defined per AGA guidelines (22, 139) and are used to define the study group. A subject who has had a flexible sigmoidoscopy is eligible. An incomplete colonoscopy is one where the prep was considered “poor” or more than 15% obscured (see SOP, Appendix E) or the entire colon could not be visualized or the scope did not reach the cecum (unless an obstructing mass was the reason the scope didn’t reach the cecum). Subjects having a repeat colonoscopy due to a previous “incomplete” colonoscopy are eligible if they otherwise meet inclusion/exclusion criteria because incomplete colonoscopies are not a “full colonoscopy”.
5.4.2Minimum Requirements for Subject Enrollment
a.Two FIT tests shipped properly per SOPs (within tolerance)
b.Stool samples shipped properly per SOPs (within tolerance)
c.Blood: minimum 18 aliquots of serum, 18 aliquots of plasma, and 2 huffy coats processed per SOPs
d.Complete colonoscopy to cecum with good or better bowel preparation (per colonoscopy SOPs) or an obstructing mass prohibiting insertion to cecum
e.All data forms
f.Four 5 ml vials of urine
5.4.3Enrolled Subject
An enrolled subject is one that has signed the informed consent, is eligible based on inclusion and exclusion criteria at the time of consent (section 5.2) AND has the minimum specimens required (5.4.2). Replacement samples or additional visits before the screening colonoscopy are options to meet the minimum requirements to enroll a subject. Once a subject meets the inclusion and exclusion criteria and provides specimens within the 16- week window, the qualifying colonoscopy for study purposes is the first one that is complete (to cecum) with a good or better preparation (defined as less than 15% of mucosa obscured). An otherwise eligible subject may need a repeat colonoscopy due to poor prep, poor sedation , or some other technical or logistical issue. These subjects would be considered enrolled as long as the colonoscopy is done within 16 weeks of original specimen collection. Enrolled subjects are listed as “pending” in VSIMS until confirmed. (See 5.4.5) or deemed ineligible (see 5.4.6).
5.4.4Protocol Deviations
Subjects who do not meet the minimum requirements (5.4.2), do not have a complete colonoscopy or have to provide replacement samples will not be reported as protocol deviations.
5.4.5Evaluable Subjects
Once an enrolled subject has completed their colonoscopy, and the recruiting site has pathology and colonoscopy reports, the site should run “Confirm Eligibility” in VSIMS. The “Confirm Eligibility” function will verify that the subject met the inclusion/exclusion criteria, provided the required samples and data (5.4.2), and count the subject in a final group or bin based on the colonoscopy results (including no colonoscopy). Evaluable subject’s samples and data will be used for analysis or building a reference set.
Bin #1 - Colorectal Cancer
Bin #2 - Carcinoma in Situ
Bin #3 - Adenoma with High-Grade Dysplasia
Bin #4 - Advanced Adenoma
Bin #5 - Adenoma
Bin #6 - Hyperplastic Polyps
Bin #7 - Polyp of Other & Unknown Types
Bin #11 - Normal Colon
12
5.4.6Screen Failures, Not Eligible and Unevaluable Subjects
Subjects who are approached to participate via a face-to-face visit and do not meet the eligibility criteria in section 5.2 are “screen failures”. These subjects will not be entered in VSIMS, should not be issued a PID, and will not receive payment for samples ($25).
Screen Failures will not count as accruals.
Subjects who sign the informed consent, but end up not meeting the eligibility criteria in section 5.2 (with samples already collected) will be “ineligible”. Subjects who meet the inclusion/exclusion criteria but do not provide stool and blood will be labeled ineligible. An “ineligible” CRF will be completed. Ineligible subjects will be entered in VSISMs and may receive payment for stool samples if provided before determined ineligible. Ineligible subjects will not count as accruals toward the total 9000 subjects.
Subjects who are eligible, sign the consent form, and then do not meet the minimum requirements for subject enrollment (section 5.4.2) are unevaluable. Subjects that provide specimens but do not have a colonoscopy or subjects that have a colonoscopy with poor prep, poor sedation or an otherwise incomplete colonoscopy are considered unevaluable. The site should run “Confirm Eligibility”. Unevaluable subjects will be entered in VSIMS and may receive payment for stool samples if provided. Unevaluable subjects will count as accruals toward the total 9000 subjects.
Bin #8 - Uncategorized Colonoscopy
Bin #9 - Incomplete Colonoscopy
Bin #10 - No Colonoscopy
5.4.7Off-Study
A subject is off-study when the data, food frequency questionnaire, blood, urine and stool samples (including both FIT tests) have been obtained, properly processed and delivered to, the Central Laboratory at the University of Michigan/DKFZ, a colonoscopy has been completed, eligibility confirmed and the one year follow up contact has been conducted.
Data collection will continue on subjects that have findings of cancer or require surgical excision of lesions (i.e. adenomas) in order to obtain staging, treatment, and outcomes relevant to the use of the biomarkers, and these subjects will not be off study until that data collection is complete. Adverse events or serious adverse events will not be reported for subjects remaining on study between the completion of their baseline visit and going “off study” as this is a minimal risk, non-interventional study (section 9.6).
5.5Biological Sample and Data Collection
5.5.1Blood Collection, Processing and Storage
Subjects will provide 50 ml of blood as defined above. Blood samples will be drawn in a specific order: 2 x 10 ml red top tubes and then 3 x 10 ml purple top tubes. Purple tops tubes must be filled to manufacturer’s level to maintain blood:EDTA ratio. Sufficient blood is needed to ensure that a minimum of 18 aliquots of serum, 18 aliquots of plasma and 2 huffy coats are collected. Additional red and purple top tubes may be collected to get the full 50 mls needed. Additional blood draws, prior to prepping for the colonoscopy may be done to get to the necessary blood volume.
The serum samples (red top tubes) will sit at room temperature for a minimum of 30 minutes (maximum of 60 minutes) to allow the clot to form, and if not processed immediately, they can be held at 4° C for a maximum of 4 hours after collection. Plasma samples (purple {aka lavender} top tubes) will be held at 4° C for a maximum of 4 hours after collection. The red top collection tubes will be centrifuged at>1,300 x g at 4° C for 20 minutes. The serum will be removed, transferred to pre-labeled polypropylene capped tubes, and frozen at - 70° C or colder. The purple top collection tubes will be centrifuged at >1,300 x g at 4° C for 10 minutes without the brake on the centrifuge. The plasma will be transferred to a 15 ml conical tube for a second centrifugation step (>1,300 x g at 4° C for 10 minutes) prior to aliquoting in pre-labeled polypropylene capped tubes, and frozen at - 70° C or colder. The huffy coat, remaining in the purple top tubes above the red blood cells, will be removed and placed into 2 pre-labeled vials, up to 1.2 mls of RNALater® (Sigma Chemical Corp, St. Louis, MO) will be added and stored at -70° C or colder. All frozen samples will be stored at - 70° C or colder at the collection site and shipped on dry ice to the CLASS labs at the University of Michigan and stored at- 70° C or colder until assayed. Detailed Standard Operating Procedures including shipping and sample handling instructions are provided in Appendix C.
13
5.5.2Urine sample collection, processing, storage
At the baseline visit, subjects will be asked to provide a urine sample of at least 25 mls. The urine specimen will be stabilized with IM EDTA, and held at 4°C for up to 4 hours until aliquoted. The urine will be aliquoted and stored frozen at -70° C or colder. All frozen samples will be stored at - 70° C or colder at the collection site and shipped on dry ice to the CLASS labs at the University of Michigan and stored at - 70° C or colder until assayed. Detailed Standard Operating Procedures including shipping and sample handling instructions are provided in Appendix G.
5.5.3FIT Analysis
Subjects will be provided with a standard collection kit including detailed instructions on how to complete the FIT sampling (Appendix D). The first FIT tube will be shipped inside the same shipping container with the stool sample (see 5.3.5). The second FIT tube will be mailed (pre-paid) to the University of Michigan or DKFZ at room temperature in the manufacturer’s DOT-compliant envelope. The test will be analyzed at the Central Laboratory at the UM or DKFZ using analytic equipment provided by Eiken Chemical Company. (OC-SENSOR Xxxxx).
5.5.4Stool Sample Collection and Handling
Subjects will be asked to collect their stool in the collection bucket (hat) provided. Subjects will be given detailed instructions and complete kits to collect the stool samples at home.
They will prepare a FIT tube (FIT #1) from the stool sample. Subjects will also collect scoops of stool into a container with an EDTA-based buffer (“buffered stool”) and additional scoops of stool into tubes provided to be sent on ice packs (“native stool”) The subjects will then package both the stool and the FIT for shipping per provided instructions. The US and Canadian subjects will ship the stool sample to the Central Laboratory at the University of Michigan using pre-paid DOT (Department of Transportation)-compliant packaging. German subjects will send their stool samples to the German Cancer Center (DKFZ) (Xx. X. Xxxxxxx). Buffered stool samples will be homogenized and frozen in four 5 ml aliquots at -70° C or colder for batch shipment to the analytical labs. The native stool will be placed at - 70° C or colder upon receipt.
5.5.5Follow up
The CRA will contact the subject via phone or letter or email about one year (window 11- 14 months) after their qualifying colonoscopy for additional follow up data including changes in family history of cancers, significant personal medical events such as hospitalizations or new medical diagnoses, and any diagnosis or treatments for cancer or dysplastic lesions (e.g. adenomas). Data will be collected on medical record review forms and follow up data forms (Appendix A).
5.5.6Medical Records Documentation
Medical records will be reviewed to collect information regarding the results of the procedures, pathology analysis, surgery, treatment, history, or outcomes and documented in the CRFs. The medical records will serve as the source documents and will be maintained at the site enrolling the subject. Redacted copies (identifiers blocked out) of colonoscopy reports and pathology reports may need to be sent to the University of Michigan for review. Medical records and/or source documents may be reviewed at the site during audits or monitoring visits.
5.6Disclosure of results to subjects
Subjects will be informed as part of the consent process that neither they nor their health care providers will receive any subject-specific results from participation in this study including results of FIT, stool or blood sample assay results. Subjects and their health care providers will be furnished with published data (abstracts, published manuscripts) upon request.
5.7Biomarker Analytical Approach
5.7.1Sample Shipment to Analytical Sites
Samples will be shipped in batches to analytical sites. Shipment date and time will be recorded in shipping logs in VSIMS. Date and time of receipt will be recorded at the analytical laboratory. Laboratory staff will be blinded to all subject data except specimen ID and relevant handling or processing information . No diagnostic or additional demographic data will be provided. The analytical laboratories will not have access to the database, and will not be able to link the bar code to a specific PID.
14
5.7.2Analytical Performance and Reporting Standard
The laboratory will have 12 weeks (3 months) from the date of sample receipt to complete the analytical task and report the data to the DMCC. The data will be reviewed for quality control by the biostatistician (DMCC). If there are concerns regardin g variance of the data, an on-site visit will occur to review the methods of assay quality control and data manipulation.
5.8Data Collection, Management and Monitoring
Data will be collected, managed and monitored through the EDRN supported VSIMS system of the Data Management and Coordinating Center (DMCC). This system is a fully featured, Good Computing Practice compliant (secure, audit trail, daily backup) database system with biosample tracking capability. Data will be entered via a Web-fronted interface at each collaborating clinical site. The data will be subject to internal and external audits. The DMCC and GLNE Prevention Research Base at the UM will organize and implement on site audit procedures. Biosample tracking ·will be accomplished using a bar code reader and the VSIMS system in real time for each step in sample management.
Procedures | Baseline1 | Stool collection2 | Colonoscopy | Follow up5 |
Informed Consent | X |
|
|
|
Study Documentation CRF4 | X |
|
|
|
General Information CDE4 | X |
|
|
|
Medical History CDE4 | X |
|
|
|
Concomitant meds CRF | X |
|
|
|
Hormone Replacement TherapyCRF | X* |
|
|
|
Colonoscopy CRF |
|
| X |
|
Surgery Report CRF |
|
| X* | X* |
CRC treatment CRF |
|
|
| X* |
Ineligible CRF | X* |
|
|
|
Food Frequency Data Collection | X |
|
|
|
Blood Collection | X |
|
|
|
Urine Collection | X |
|
|
|
FITxl |
| X |
|
|
Protocol deviation CRF |
| X* | X* | X* |
Stool Collection |
| X |
|
|
Fixed Tissue3 |
|
|
| X3 |
One Year Follow up |
|
|
| X5 |
Subject Expense Payment |
| X |
|
|
Protocol deviation CRF |
| X | X | X |
Study Termination CRF |
|
|
| X |
1Visit prior to colonoscopy procedure; subject should not have started the colon preparation procedures.
2Stool collection any time after first visit and subject returns home with kits. All samples collected prior to beginning colon preparation procedures
3Fixed slides are obtained for pathology review.
4CDE=Common Data Elements; CRF=Case Report Fonn Note: CDEs and CRFs are in Appendix A
5To be completed 12 months after the completion of the colonoscopy
* Only if needed/applicable
15
7.1Vimentin methylation
This assay will be performed at an EDRN-designated BRL according to previously published methods described in the background. The assay will be run both qualitatively and quantitatively for presence of and quantity of methylated vimentin gene by the University of Maryland BRL (Pl Xxxxx Xxxxx). The vimentin methylation assay will be performed blinded without knowledge of clinical source or results of other assays.
7.2Fecal immunochemical Test (FIT)
The QC-SENSOR product will be used according to manufacturer’s instructions. The threshold for a positive test is 100 ng/ml. The Central Laboratory at UM and DKFZ will process the samples using equipment provided by Eiken Chemical Company. Technicians will undergo tutorial and quality assessment with Eiken Chemical Company support technicians prior to study launch. A quantitative result will be generated and recorded and uploaded into VSIMS by the DMCC. If either stool result is above the recommended cut off, that subject will be called positive.
The analytically validated ELISA method described in the preliminary data will be transferred to an EDRN Biomarker Reference Laboratory. Serum aliquots will be provided to the analytical sites in a blinded fashion. The Xxxxxxxxx laboratory will assay 20% of the samples to ensure quality control. All of the samples will be assayed by the UCLA Biomarker Reference Laboratory. Galectin-3 Ligand assay result is a continuous variable. To facilitate comparison, a threshold corresponding to the specificity of vimentin methylation, estimated from controls in this study that do not have SR,N” from colonoscopy, will be used.
7.4Circulating methylated genes BCATl/IKZFl (Clinical Genomics)
A Good Laboratory Practice validated bisulfite PCR assay developed by Clinical Genomics will be used for this assay. Clinical Genomics will perform this assay on blinded samples at their laboratory facility in Rutherford, NJ. Clinical Genomics is not responsible for analysis of any other biomarkers other than their BCATl/IKZFl product.
7.5Hypornethylated LINEl from circulating cell free DNA (VolitionRx)
A Good Laboratory Practice validated assay developed by VolitionRx will be used for this assay. VolitionRx will perform this assay on blinded samples at their laboratory facility in Namur, Belgium. Volition is not responsible for analysis of any other biomarkers other than their hypomethylated LINE I assay.
8.0DATA ANALYSIS PLAN, SAMPLE SIZE JUSTIFICATION, AND STATISTICAL POWER
Aim 1: This aim proposes to estimate the sensitivity and specificity for 1) colorectal adenocarcinoma or 2) screen relevant neoplasms (high grade dysplasia or adenoma with 2:25% villous histologic features or adenoma measuring 2:1 cm in the greatest dimension or sessile serrated polyps measuring 2:1 cm in diameter) of the following individual colorectal neoplasia early detection biomarkers using colonoscopy as the gold standard:
stool vimentin methylation
serum galectin-3 ligand
fecal immunochemical tests (FIT)
Circulating methylated genes BCATl/JKZFI (Clinical Genomics)
Hypomethylated LINE! from circulating cell free DNA (Volition)
Other currently unspecified biomarkers
For each of the individual biomarkers, we will first verify through a receiver operating curve (ROC) analysis the previously established thresholds for an optimum sensitivity and specificity. For ties in area ofROC, we shall choose the cut-off based on the highest percent agreement.
From this point on, we will treat the performance of the markers as a dichotomy. We will calculate accuracy summaries such as sensitivity, specificity, predictive values, as well as the likelihood ratios for each individual marker along with the associated confidence intervals.
16
Aim 2 (primary objective): Three potential scenarios of possible primary hypotheses and corresponding plans for data analysis and statistical power are presented. The choice of primary hypothesis will be finalized prior to data analysis based on state of the art information about candidate biomarkers and clinical practice at that time.
x.Xx determine if a blood based panel (for example, serum galectin-3 ligand, CEA, methylated genes BCATl/lKZFl, Hypomethylated LINEl from circulating cell free DNA), at the same sensitivity of that for fecal immunochemical testing (FIT) for the detection of colorectal adenocarcinoma, has a specificity greater than 0.55 with an anticipated specificity 2’.: 0.70.
x.Xx determine if stool vimentin methylation and the blood based panel (serum galectin-3 ligand, CEA, methylated genes BCATl/IKZFl, Hypomethylated LINEl from circulating cell free DNA) when combined with fecal immunochemical testing (FIT) will significantly improve the sensitivity of FIT for the detection of colorectal adenocarcinoma, and maintain specificity greater than 0.80.
x.Xx determine if stool vimentin methylation, the blood based panel (serum galectin-3 ligand, CEA, methylated genes BCATl/IKZFl, Hypomethylated LINEl from circulating cell free DNA), when combined will improve the detection of colorectal adenocarcinoma: at sensitivity 2’.:0.98 it will have a specificity significantly greater than 0.55.
The statistical power analysis assumes 71 CRC cases (63 from the additional 9,000 subjects plus 10 CRC cases already recruited less 2 CRC cases in the surveillance sub-cohort), and>13,000 non-CRC controls (assuming ~12,000 non-SRN controls and ~1,300 SRNs).
Example a: A blood based panel (serum ga]ectin-3 ligand, or combined with other blood based biomarker if necessary) will be defined and locked-down prior to data analysis. At a cutoff with the same sensitivity as FIT (assumed here to be 0.75 but the value will be estimated from the study data) the specificity for non-SRN controls and its 95% C.I. will be calculated. The I-sided hypothesis that this specificity is significantly higher than 0.55 (Ho) will be tested with an anticipated specificity 2:: 0.70 (H1), using the kernel method described by Bantis and Feng (141). The kernel ROC estimate has been proven to have smaller mean square error than that of the empirical ROC estimate (142).
Statistical power will be >85% or >90% with an anticipated specificity 2’.: 0.70 or 2’.: 0.75 respectively. Preliminary data from GLNE investigator Xxxxxx Xxxxxxxxx supports the performance assumptions made. The blood based panel used Galectin 3 ligand, methylated genes BCATl/IKZFl, Hypomethylated LINEl from circulating cell free DNA, CEA with four different modeling approaches using GLNE data (94 normals, 50 small adenomas, 100 CRCs, and 51 advanced adenomas). The specificity at 75-80% sensitivity is >70% for both negative colonoscopy group and for negative colonoscopy plus small adenoma group.
Example b: To determine if stool vimentin methylation and the blood based panel (serum galectin-3 ligand , CEA, methylated genes BCATl/IKZFl, Hypomethylated LINEl from circulating cell free DNA) will significantly improve the sensitivity of fecal immunochemical testing (FIT) for the detection of colorectal adenocarcinoma, and maintain specificity greater than 0.80. From other training samples, the cutoff points and the combination rules of vimentin methylation and blood based panel will be detem1ined based on their ability to detect FIT negative colon cancer and maintain high specificity, then combined with FIT either by an “OR” rule or a linear combination. This decision rule will be locked-down prior to GLNE10 protocol data analysis. The difference of the sensitivities of this decision rule and FIT and the I-sided 95% confidence interval of this difference will be calculated from 10,000 bootstrap samples to accommodate the dependence between two tests. The null hypothesis will be rejected if the lower bound of this confidence interval is above zero.
The statistical power will be >0.83 if the true difference in sensitivity is 2’:.0.16 (e . g. 0.75 for FIT, 0.91 for the new test) under the conservative assumption of independence of the two tests. The actual power will be larger as these two tests are expected to be positively correlated.
Example c: To determine if stool vimentin methylation, the blood based panel (serum galectin-3 ligand, CEA, methylated genes BCAT I /IKZF l , Hypomethylated LINEl from circulating cell free DNA), when combined will increase the detection of colorectal adenocarcinoma: at sensitivity 2’: 0.98 it will have a specificity significantly greater than 0.55, with an anticipated specificity 2’: 0.79. This has significant clinical value as it has potential to spare more than 55% people in US from colonoscopy screening, and improve screening rate for those who do not want to have colonoscopy as first-line screening modality. From other training samples, a panel will be built to achieve 2’: 0.98 in sensitivity in detecting colorectal adenocarcinoma while maintaining specificity 2’: 0.79 for subjects without SRN lesions. This is feasible if other markers can pick up majority of FIT negative CRCs without reducing specificity by more than 15%. The combination rule will be defined and locked-down prior to GLNEl O protocol data analysis. To test the study hypothesis, at cutoff corresponding to 0.98 sensitivity, the specificity of this decision rule, its I sided 95% C.I., and the I-sided hypothesis that this specificity is significantly higher than 0.55 (Ho) will be tested using the method using the kernel method described by Bantis and Feng (141). Statistical power is > 0.90 if the true specificity 2: 0.79. The 1-sided 95% C.I. for sensitivity will also be reported.
17
Secondary objective: While the study will be powered for the primary objective, we shall also carry out a similar assessment of potential utility of a clinical test for SRN.
Aim 3 To construct a combined early detection biomarker panel using the above individual biomarkers (stool vimentin methylation, serum galectin-3 ligand, FIT, the Exact Sciences stool DNA panel, circulating methylated genes BCATl /IKZFl , hypomethylated LINEI from circulating cell free DNA), and describe its performance for 1) colorectal adenocarcinoma and for 2) screen relevant neoplasms.
After the primary hypothesis (Aim 2) has been finalized, the examples described in Aim 2 and are not chosen as the primary hypotheses will be tested in Aim 3 as they are all clinically relevant hypotheses. The data analyses and their statistical power for these hypotheses have been described under Aim 2 above and so are not repeated here.
In addition, new panels could be constructed using the trial data as training set but these panels will need to be validated on other independent cohorts.
Aim 4
To establish an archive of appropriately preserved stool, serum, plasma and DNA human biospecimens to be used by EDRN-approved investigators for future validation and biomarker discovery research.
8.1.1Secondary Analyses for Screen Relevant Neoplasias
Similar analyses as described above will be performed for the secondary endpoint (SRN). The minimally acceptable performance used for setting null hypotheses will differ: sensitivity=0.25 for secondary specific aim 2, and sensitivity=0.45 for secondary specific aim 3.
8.1.2Secondary Analyses for projecting biomarker performance in US population
Since our enrollment is enriched with older subjects, in Aim la we will also report age and gender-standardized accuracy summaries that is calculated with an inverse probability weighted method in order to adequately reflect the performance of biomarkers in the US screening population which may have a different age distribution from that of our study population. The ratio of observed proportion within each age by gender stratum versus the corresponding proportion in the US population as reported by the Census data will be used as the weights for subjects in the stratum.
Note that the weighted analyses may provide reasonable projections for biomarkers’ performance in the US population, however we will treat this as a secondary analysis and therefore our power calculation is based on unweighted analyses from the observed sample.
8.1.3Additional Secondary Analyses
The cohort will be characterized in terms of demographics and epidemiological variables such as gender, education status, intake of dietary micro and macronutrients, hormone replacement therapy, alcohol intake and smoking. We will assess the relationship of each of these factors individually to each marker under evaluation and risk of colorectal cancer in multiple regression models. For factors that modulate the relationship between biomarkers and clinical endpoints, we will further evaluate their effects on the accuracy parameters of the marker, by evaluating for example the covariate-specific ROC curves. We will also test directly covariate effects on the accuracy of markers in order to identify subpopulations for which markers are mostly effective.
8.1.4Inclusion of New Biomarkers Discovered by EDRN Investigators over the Next Two Years
The design of this project including the collection of serum, DNA and tissue samples permit the inclusion of new EDR._ discovered biomarkers into this panel. Should EDR.i”“\J” investigators provide sufficient preliminary data to justify inclusion in this panel; new biomarkers will be included in the validation program using the procedures described above.
18
9.1Strategies to Ensure Completion of Milestones
Milestones set up and regularly reviewed: Milestones are set on a quarterly schedule and managed by the principal investigator.
Conference calls: a) Investigator calls: All GLNE investigators communicate every other week by scheduled telephone conference call organized and chaired by the PI, Xx. Xxxxxxx (11 AM to 12 Noon, Thursdays). b) Coordinator calls: All GLNE research coordinators and support staff communicate once per month by scheduled telephone conference call organized and chaired by the Project Manager, Xx. Xxxx Xxxxxx and the DMCC.
Data and Safety Monitoring: The University of Michigan Cancer Center Prevention Program’ s Data Safety and Monitoring Committee meets monthly and reviews progress towards milestones. Accrual, endpoints, toxicity, and strategies to ensure goals are met reviewed by this committee. Minutes are forwarded to the supervising IRB (IRBMED) and to relevant regulatory agencies.
9.2Timeline for Completing GLNE 010
Table 2, a Xxxxx diagram, outlines our milestones for the proposed CRC early detection biomarker trial.
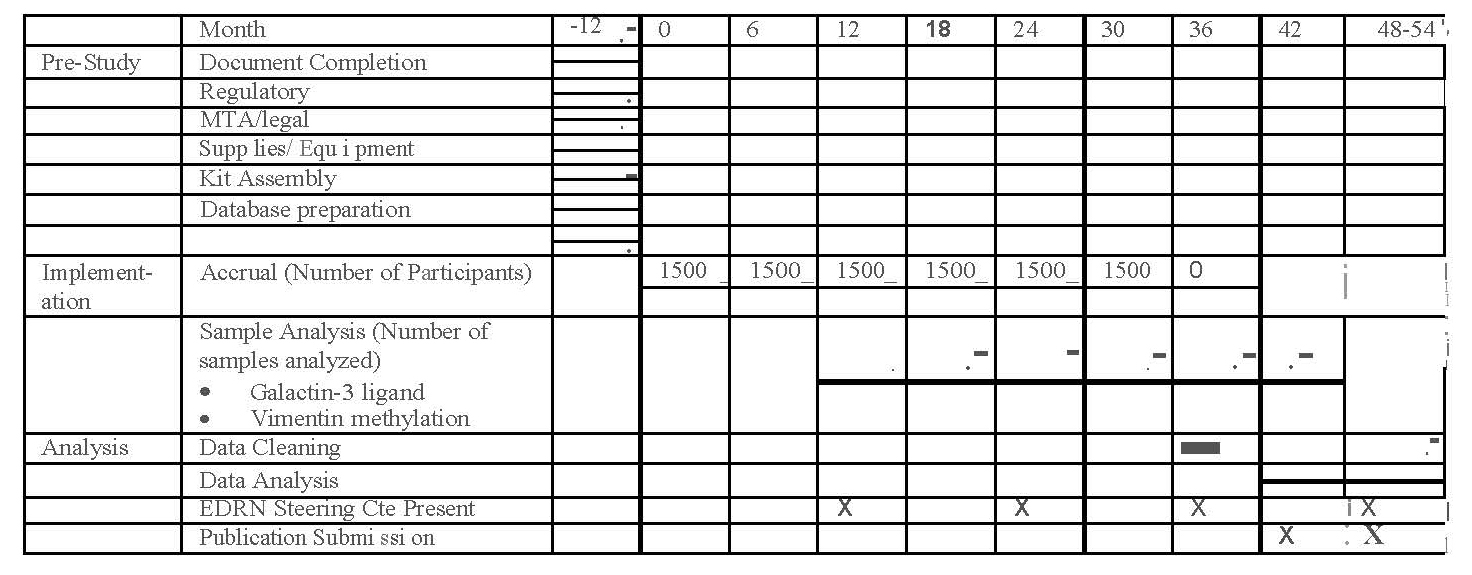
Table 2: GLNE 010 (Colon Biomarker Validation Trial) Milestones
We have broken down this trial into the re-organizational phase (Months -6-0) that began on April 6, 2016 and will be completed on April 1, 2017. Accrual phase beginning April 1, 2017, will last 3.5 years with data assay and data analysis being complete in Years 5. Milestones are outlined below and add detail to the events depicted in Table 2. We estimate 250 participants/month accrual at all sites (EDRN, Alliance/NCORP, and Germany, see Table 3).
Center | Accrue/mo | Pl | %Minority |
Case Western | 2 | Coooer | 40% |
Columbia | 2 | Kastrinos | 80% |
Xxxx Xxxxxx | 7 | Xxxxxx | 10% |
MD Xxxxxxxx | 8 | Xxxxxxxxx | 20% |
Minnesota | 30 | Xxxxx/Church | 3% |
St Michaels/Toronto | 15 | Marcon | 3% |
Univ IL Chicaqo | 1 | Xxxxxxx | 55% |
Univ Michigan | 12 | Xxxxxxx | 3% |
Univ North Carolina | 13 | Xxxxxxxx | 1% |
Univ Washington | 11 | Gradv | 1% |
Alliance/NCORP-Mavo | 61 | Xxxxxxxx | 3% |
Total USA | 162 |
|
|
Heidelbera, Germanv | 95 | H Xxxxxxx | 0% |
Total USA-Germany | 257 |
|
|
Table 3 (Left): Prior accrual experience, GLNE USA, limited to age .!:65 yrs.
DKFZ Germany documented accrual, all age groups over 7 years, BliTz (personal communication, X. Xxxxxxx).
19
9.3Endpoint Event Justification and Milestones
9.3.1Required Endpoints
In order to successfully complete this project a minimum of 70 invasive colorectal neoplasms must be detected. To ensure we reach this endpoint, we revised the protocol to 1. Expand the trial to include Germany; 2. Limit recruitment in USA sites to participants 60 years or older undergoing first colonoscopy.
9.3.2Justification for Requiring only Invasive Colorectal Neoplasms
First, recent as well as older large validation trials have found that dysplastic adenomas as endpoints degrade biomarker detection accuracy (102, 103). For example, in the recently published Exact Sciences stool DNA panel, sensitivity for invasive colorectal neoplasms only was 92% which reduced to 84% when high grade dysplastic adenomas were included with invasive colorectal neoplasms (103). Second, a strategy which allows high grade dysplasia in adenomas as primary endpoints reduces the likelihood of successful identification of early detection biomarkers for colorectal neoplastic disease. We run the risk oflosing biomarkers to failure requiring classifier performance of single markers or panels that might be useful for the detection of curable invasive colorectal neoplasms despite poorer performance for detection of high grade but non-invasive neoplasms. We increase barriers for success to the very biomarkers that the EDRN exists to discover and validate. Third, inclusion of high grade dysplasia in adenomas with invasive colorectal neoplasms degrades the usefulness of the repository samples generated by GLNE 010.
There will be insufficient invasive colorectal neoplasm events to use for large cross sectional validation of future biomarkers. Our only other resource for invasive colorectal neoplasms is a cross sectional reference set that is not PRoBE compliant.
9.3.3Strategies to Ensure Sufficient Invasive Colorectal Cancer Events
Expand GLNE to Germany: The rapid expansion of colonoscopic screening in the USA to younger age groups (ages 50-59) (Table 4) with high adherence to colonoscopic screening guidelines (61%), while reducing incidence and presumably mortality from invasive colorectal neoplasms may be reducing the numbers of screen detected invasive colorectal neoplasms in USA screening trials such as GLNE 010. In GLNE 010, the detected rate of screen relevant (“advanced”) adenomas of 13% exceeds the expected rates in recently published biomarker trials using screening populations [Xxxxxxxxx, 2014 #5685;Xxxxxxxxx, 2014 #5684;Imperiale, 2014 #5977;Church, 2014 #4704]. In Germany, colonoscopic screening is provided as a benefit to the population, but with much lower colonoscopic screening adherence rates (16%) (25) as opposed to 61% adherence in the USA (19). The higher invasive colorectal neoplasm case proportion of 0.7% in a large ongoing prospective screening trial (Table 4) makes a primary colorectal neoplasm endpoint feasible.
Age | Exact | PRESEPT | XxxXx | XXXX |
00-00 | 29% | 35% | 36% | 42% |
60-64 | 8% | 27% | 23% | 18% |
65-69 | 37% | 21% | 25% | 21% |
70-74 | 17% | 11% | 13% | 21%* |
?:.75 | 9% | 6% | 4% | --- |
CRC Event | 0.7% | 0.8% | 0.7% | 0.2% |
HGD Event | 0.4% | 0.7% | 0.7% | 0.4% |
Table 4: Enrollment Ages, Recent Large Cross Section CRC Screening Trials.
Exact Sciences (2014, (1)), PRESEPT (2014, (2)), BliTz
(ongoing, Heidelberg, X. Xxxxxxx, Personal Communication.)
Enrich North America Risk to Bring Endpoint Event Percentages to the Level of Other Trials: The incidence of invasive colorectal neoplasm increases with age, particularly at ?:.60 yrs. The recent published PRESEPT and Exact Sciences trial enriched their screening populations to increase invasive colorectal neoplasm event rates. PRESEPT restricted enrollment to first colonoscopic procedures. PRESEPT did not limit age of enrollment, two thirds of participants were ?:.60 yrs. Exact Sciences required two thirds of enrolled participants be ?:.65 yrs, but permitted prior colonoscopic screens ?:.9 yrs. BliTz has no age limits or prior endoscopy limits, but the low screening adherence rate in Germany suggests that the population represents first colonoscopic screening procedures.
20
9.4.1Endpoint Monitoring
As part of weekly accrual monitoring, endpoints events (invasive cancers, high grade dysplasia, screen relevant neoplasia, and adenoma) will be reported.
9.4.2Monitoring Primary Endpoint Events
The primary endpoint, invasive colorectal cancer, will be monitored weekly.
9.4.3Primary Endpoint Expectations
At an expected accrual rate of 1,500 per 6 months, we expect 10 invasive cancers to be detected every 6 months. Because events do not occur at regularly spaced intervals, but rather as randomly distributed events, to identify potential problems in meeting our primary endpoint goal we need a sufficient number of evaluable accruals to determine whether the current enrollment strategy will meet the accrual goal of at least 70 invasive neoplasms.
For these reasons, we will wait until 1,000 subjects have been enrolled to assess whether our case event rates will be sufficient to meet the current primary study endpoint.
9.4.4Monitoring to Ensure Sufficient Cancer Endpoints
The following procedure will be followed in collaboration with the DMCC: Review every 1,000 evaluable subjects through the course of the project with the expectation that 7±1 new invasive neoplasm cases will be detected for every 1,000 evaluable subjects. If <7 new invasive neoplasm cases are enrolled for every 1,000 evaluable subjects , the project will be reviewed by statistical consultants and coinvestigators and revision of study goals and design by allowing FIT positive screens.
9.5.1Authority
The University of Michigan Prevention Research Base Data Safety and Monitoring Committee (DSMC) reviews, makes recommendations, and acts on the following:
a.All protocols being run through the GLNE EDRN will be monitored by the DSMC
b.Progress towards completion of the study-recruitment and retention of study participants
c.Evaluation of interim new information
d.Evaluation of toxicity events including reporting of adverse events, if applicable
e.Timeliness of data
f.Quality of data
g.Ethical conduct of research
The DSMC is empowered with the authority to recommend a study be suspended or terminated based upon concerns in any of the above areas ofreview. Monitoring also considers factors external to the study, such as scientific or therapeutic developments that may have an impact on the safety of the participants or the ethics of the study.
Recommendations that emanate from monitoring activities are reviewed by the principal investigator and addressed.
9.5.2Composition
The current UM Prevention Research Base Data and Safety Monitoring Committee is Chaired by one of the faculty members present at the meeting, usually the most senior member who is not a principal investigator on studies being discussed. Membership includes faculty members from Gastroenterology, Family Medicine, and Hematology/Oncology. At least 3 faculty members must be present to have quorum. If the DSMC cannot meet face-to-face, a conference call is acceptable.
21
9.5.3Meeting Frequency
The UM Prevention Research Base DSMC meets monthly by means of regularly scheduled meetings. Prior to each meeting, the UM Prevention Research Base Clinical Research Associate distributes a standard summary report detailing accrual, biomarker modulations data, new publications or presentations relevant to the ongoing project, quality control audit information, any ethical concerns, patient-subject complaints and adverse events or serious adverse events of all prevention protocols.
9.5.4Recommendations and Reporting
Recommendations for action are sent to the Principal Investigator. The Principal Investigator is responsible for reviewing and if necessary, implementing DSMC recommendations.
9.6.1Definition
An adverse event (AE) is any condition, which appears or worsens after the participant is enrolled in an investigational study. For this minimal risk, sample collection study, we provide a definition of what would be considered related to the study participation.
9.6.2AE Information
No adverse events are expected, as there is no intervention for this study. Any adverse events related (as judged by the site PI, overall PI or DSMC) to the subject’s participation (sample or data collection) in this study will be forwarded to the data coordinating center and reported to regulatory bodies per study-specific guidelines. Adverse events or serious adverse events will not be reported for subjects remaining on study between the completion of their baseline visit and going off-study as this is a minimal risk, non-interventional study. Subjects could be considered on study for over a year and have adverse events completely unrelated to study participation. Examples of related adverse events that could be reported could include problems with the blood draw (bruising, fainting), loss of confidentiality of data, or lost samples. Examples of events that would not be reported would include any complications from the colonoscopy or events related to other medical conditions like colon cancer or diabetes.
9.6.3Serious Adverse Events
Some percentage of participants will have colorectal cancer identified during colonoscopy by study design, and deaths due to disease progression or serious adverse events due to cancer treatment are expected. The only procedures that are part of this study are blood, urine and stool collection, so it is unlikely that any deaths or hospitalizations will be related to the sample collection in this study. Only Serious Adverse Events that are deemed to be directly related to a study procedure (sample collection) by the DSMC will be reported to any regulatory body.
A serious adverse event is defined (by ICH Guideline E2A and Fed. Reg. 62, Oct. 7, 1997) as an event, occurring at any dose, which meets any of the following criteria:
Results in death
Is immediately life threatening
Requires inpatient hospitalization or prolongation of existing hospitalization
Results in persistent or significant disability/incapacity
Is a congenital anomaly/birth defect
In addition, events that may not meet these criteria, but which the investigator finds very unusual and/or potentially serious, will be reported in the same manner.
22
Institutional collaborators will enter IRB information into the secure database, including IRB approval date, expiration date, and document versions. Subject registrations will not be allowed without IRB approval. The DMCC will assign the participant ID number (PID).
No exceptions to eligibility requirements will be permitted.
10.2Timeliness
In collaboration with the DMCC, a data expectation system will be developed. Detailed instructions are provided in the operations manual. For sampie shipments, data from the shipping and receiving laboratory describing the date of sample shipping and sample receipt are captured in the database.
10.3Completeness and Accuracy
The DMCC will assure the completeness of the data by writing data entry programs that will not allow for empty fields whenever possible. The accuracy of the data will be checked by identifying appropriate parameters allowed to be entered in a given data field. Periodic reviews of the paper forms and the database data will be conducted by the lead CRA and a Site Monitor from the Coordinating Center.
10.4Accuracy--Revisions and Corrections
All corrections to paper study documents will be initialed and dated. If computer-readable data is corrected by replacement of a data set, the replaced version of the data set will be retained in an archive. The collection of these auxiliary data sets represents an audit trail of corrections to the database.
10.5On Site Data Audits
All consortium sites will be subject to periodic on-site audits. The objective of the on-site audit will be to conduct a general review of a random sample of registered subjects from the selected protocol to assess overall protocol adherence with respect to subject eligibility, appropriate procedure for informed consent, registration process, general protocol adherence, sample shipment process, follow-up and off-study process.
An On-Site Audit checklist will be developed which will contain all of the essential elements of an On-Site audit. Each of the essential elements will be reviewed and discussed with the clinical site. The Checklist will be signed by the auditors and retained at the DMCC.
In preparation for a site audit, the study statistician will select the subjects for review using a randomized selection procedure. Other cases may also be selected at the discretion of the audit team. A minimum of 10% of the subjects accrued since the last audit will be reviewed for the first year. The number of subjects to be audited for the subsequent year will be determined based on findings of the audits from prior years in order to have sufficient power to identify important issues. The on-site audit team will audit additional unannounced cases. The consortium site investigator and research coordinator will be notified of the impending audit at least 3 weeks in advance. All data and material pertinent to the subject will be reviewed including eligibility criteria, informed consent, and sample shipment logs. All informed consent documents for all subjects may be reviewed at the on-site audit. At the audit, the data from the DMCC will be compared to the original data (source documents and/or CRFs). On-site audit staff will review the documentation of IRB approvals, for each audited protocol, any amendments or adverse events, and consent forms.
Based on the findings of the audit, a follow-up schedule will be defined. A report of the audit will be written and emailed or faxed to the consortium site investigator. The site PI will have 30 days from receipt of the report to respond in writing to the DMCC directly.
The DMCC will maintain a file containing the latest version of the On-Site Audit guidelines, a listing of all consortium institutions reviewed to date, a copy of the On-Site Audit results and all correspondence for each audit conducted. These results will be reviewed by the DSMC, the DMCC, and others as needed and will be made available to the NCI.
23
10.6Sample Tracking
CLASS labs must be notified via e-mail of a shipment due to arrive so if samples are delayed or lost, tracking may be initiated by the sending site. Sample shipment packing lists generated by the database are included with shipments. The receiving site will evaluate the sample condition on arrival, scan the bar-coded samples into database, verify samples shipped match samples sent and store at appropriate conditions until shipment to analytical labs or repositories.
10.7Confidentiality
Subjects will be identified in the database by their unique PID only. Information that could identify subjects, such as name, address, or medical record number will be kept only by the enrolling site and will not be supplied to the DMCC or GLNE Research Base. During an on-site audit or NCI site visit, audit staff may review medical records and other information that contains PHI, but this information will not be removed from the enrolling site. Neither the DMCC nor the research base at UM will keep copies of signed informed consent documents. No information, including copies of the informed consent unless required by the institution, obtained during the study will be placed in a subject’s medical record.
10.8Security
All subject files will be stored under lock and key at all times. All computer systems will be password-protected against intrusion; all network-based communications between sites of confidential information are encrypted. An on-going computer-virus-protection program is available and used, maintained, and audited on all computers and pathways into the system, including good practice policies, screening of data files, executable software, diskettes, text macros, downloads, and other concerns as they arise. The DMCC will assist in maintaining appropriate levels of network security.
11.0ETHICAL & REGULATORY CONSIDERATIONS
11.1Institutional Review
This study must be approved by an appropriate institutional review committee as defined by Federal Regulatory Guidelines (Ref. Federal Register Vol. 46, No. 17, January 27, 1981, part 56). The protocol and informed consent form for this study must be approved in writing by the appropriate Institutional Review Board (IRB). The IRB must be from an institution that has a valid Federal Wide Assurance on file with the Office for Human Research Protections, Department of Health and Human Services. The institution must comply with regulations of the Food and Drug Administration and the Department of Health and Human Services. Changes to the protocol, consent, as well as a changes to the investigator list at each site, must also be approved by the IRB and documentation of this approval provided to the Coordinating center.
Records of the Institutional Review Board review and approval of all documents pertaining to this study must be kept on file by the investigator and are subject to OHRP, FDA or NCI inspection at any time during the study. Periodic status reports must be submitted to the Institutional Review Board at least yearly, as well as notification of completion of the study and a final report within 3 months of study completion or termination. The investigator must maintain an accurate and complete record of all submissions made to the Institutional Review Board, including a list of all reports and documents submitted.
24
12.0 REFERENCES
0.Xxxxxx TR, Xxxxxxx M, Xxxxxx-Day C, Xxxxxx XX, Burger M, Xxxxx SR, et al. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut. 2014;63:317-25. PMCID: 3913123.
2.Xxx XX, Xxxxx EG, Bent S, Xxxxx XX, Xxxxxx DA. Accuracy of fecal immunochemical tests for colorectal cancer: systematic review and meta-analysis. Xxx Intern Med. 2014;160:171. PMCID: 4189821.
3.Pepe MS, Feng Z, Xxxxx H, Bossuyt PM, Xxxxxx XX. Pivotal evaluation of the accuracy of a biomarker used for classification or prediction: standards for study design. J Natl Cancer Inst. 2008;100:1432-8.
4.Xxxxxx XX, Xxxx XX, Church TR, Xxxxxx XX, Xxxxxxx XX, Xxxxxxx XX, et al. Reducing mortality from colorectal cancer by screening for fecal occult blood. Minnesota Colon Cancer Control Study [published erratum appears in N Engl J Med 1993 Aug 26;329(9):672] [see comments]. N Engl J Med. 1993;328:1365-71.
5.Kronborg 0, Xxxxxx X, Xxxxx X, Xxxxxxxxx 0, Xxxxxxxxxxx 0. Randomised study of screening for colorectal cancer with faecal-occult-blood test. Lancet. 1996;348:1467-71.
6.Xxxxxxxxxx XX, Xxxxxxxxxxx XX, Xxxxxxxx XX, Xxxx XX, Amar SS, Xxxxxxx XX, et al. Randomised controlled trial of faecal-occult-blood screening for colorectal cancer. Lancet. 1996;348:1472-7.
7.Xxxxxxx A, Xxxxxx XX, Xxxxxxx MS, Xxxxxxx XX, Xxxx XX, Xxxxxx XX, et al. Long-term mortality after screening for colorectal cancer. N Engl J Med. 2013;369:1106-14.
8.Xxxxxx NN, Xxxxxxxxxx MA, Paszat LF, Saskin R, Xxxxxx DR, Xxxxxxxx L. Association of colonoscopy and death from colorectal cancer. Xxx Intern Med. 2009;150:1-8.
9.Singh H, Xxxxxx Z, Xxxxxx XX, Xxxxxxx XX, Mahmud SM, Bernstein CN. The reduction in colorectal cancer mortality after colonoscopy varies by site of the cancer. Gastroenterology. 2010;139:1128-37.
10.Brenner H, Chang-Claude J, Seiler CM, Rickert A, Hoffmeister M. Protection from colorectal cancer after colonoscopy: a population-based, case-control study. Ann Intern Med. 2011;154:22-30.
11.Muto T, Kamiya J, Sawada T, Konishi F, Sugihara K, Kubota Y, et al. Small flat adenoma of the large bowel with special reference to its clinicopathologic features. Dis Colon Rectum. 1985;28:847-51.
12.Rex DK, Cutler CS, Lemmel GT, Rahmani EY, Clark DW, Helper DJ, et al. Colonoscopic miss rates of adenomas determined by back-to-back colonoscopies. Gastroenterology. 1997;112:24-8.
13.Saitoh Y, Waxman I, West AB, Popnikolov NK, Gatalica Z, Watari J, et al. Prevalence and distinctive biologic features of flat colorectal adenomas in a North American population. Gastroenterology. 2001;120:1657-65.
14.Samadder NJ, Curtin K, Tuohy TM, Pappas L, Boucher K, Provenzale D, et al. Characteristics of missed or interval colorectal cancer and patient survival: a population-based study. Gastroenterology. 2014;146:950-60.
15.Barclay RL, Vicari JJ, Greenlaw RL. Effect of a time-dependent colonoscopic withdrawal protocol on adenoma detection during screening colonoscopy. Clin Gastroenterol Hepatol. 2008;6:1091-8.
16.Barclay RL, Vicari JJ, Doughty AS, Johanson JF, Greenlaw RL. Colonoscopic withdrawal times and adenoma detection during screening colonoscopy. N Engl J Med. 2006;355:2533-41.
17.Soetikno RM, Kaltenbach T, Rouse RV, Park W, Maheshwari A, Sato T, et al. Prevalence of nonpolypoid (flat and depressed) colorectal neoplasms in asymptomatic and symptomatic adults. JAMA. 2008;299:1027-35.
18.Zauber AG, Winawer SJ, O’Brien MJ, Lansdorp-Vogelaar I, van Ballegooijen M, Hankey BF, et al. Colonoscopic polypectomy and long-term prevention of colorectal-cancer deaths. N Engl J Med. 2012;366:687-96. PMCID: 3322371.
19.Salas D, Vanaclocha M, Ibanez J, Molina-Barcelo A, Hernandez V, Cubiella J, et al. Participation and detection rates by age and sex for colonoscopy versus fecal immunochemical testing in colorectal cancer screening. Cancer Causes Control. 2014;25:985-97.
20.Quintero E, Castells A, Bujanda L, Cubiella J, Salas D, Lanas A, et al. Colonoscopy versus fecal immunochemical testing in colorectal-cancer screening. N Engl J Med. 2012;366:697-706.
21.Welch HG, Black WC. Overdiagnosis in cancer. J Natl Cancer Inst. 2010;102:605-13.
22.Levin B, Lieberman DA, McFarland B, Andrews KS, Brooks D, Bond J, et al. Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: a joint guideline from the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. Gastroenterology. 2008;134:1570- 95.
23.Force USPST. Screening for colorectal cancer: U.S. Preventive Services Task Force recommendation statement.[see comment][summary for patients in Ann Intern Med. 2008 Nov 4;149(9):1-44; PMID: 18838719]. Annals of lnternal Medicine. 2008;149:627-37.
24.Institute NC. International Cancer Screening Network Bethesda, MD2014. Available from: xxxx://xxxxxxxxxxxxxxx.xxxxxx.xxx/xxxx/xxxxxxxxxx/xxxxxxxxx.xxxx.
25.Pox CP, Altenhofen L, Brenner H, Theilmeier A, Von Stillfried D, Schmiege! W. Efficacy of a nationwide screening colonoscopy program for colorectal cancer. Gastroenterology. 2012;142:1460-7 e2.
26.Centers for Disease C, Prevention. Vital signs: colorectal cancer screening test use-- United States, 2012. MMWR Morb Mortal Wkly Rep. 2013;62:881-8.
27.Surveillance E, and End Results Program. SEER Stat Fact Sheets: Colon and Rectum Cancer: Centers for Disease Control; 2014 [cited 2014]. Available from: xxxx://xxxx.xxxxxx.xxx/xxxxxxxxx/xxxx/xxXxxxxx.xxxx.
25
28.Green BB, Wang CY, Anderson ML, Chubak J, Meenan RT, Vernon SW, et al. An automated intervention with stepped increases in support to increase uptake of colorectal cancer screening: a randomized trial. Ann Intern Med. 2013;158:301-11. PMCID: 3953144.
29.Harewood GC, Wiersema MJ, Melton LJ, 3rd. A prospective, controlled assessment of factors influencing acceptance of screening colonoscopy. Am J Gastroenterol. 2002;97:3186-94.
30.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. PMCID: 4017867.
31.Hibi K, Robinson CR, Booker S, Wu L, Hamilton SR, Sidransky D, et al. Molecular detection of genetic alterations in the serum of colorectal cancer patients. Cancer Res. 1998;58:1405-7 .
32.Vlems FA, Diepstra JH, Cornelissen IM, Ligtenberg MJ, Wobbes T, Punt CJ, et al. Investigations for a multi-marker RT-PCR to improve sensitivity of disseminated tumor cell detection. Anticancer Res. 2003;23:179-86.
33.Yamaguchi K, Takagi Y, Aoki S, Futamura M, Saji S. Significant detection of circulating cancer cells in the blood by reverse transcriptase-polymerase chain reaction during colorectal cancer resection. Ann Surg. 2000;232:58-65.
34.Zou H, Yu B, Wang Z, Sun J, Cang H, Gao F, et al. Detection of aberrant pl6 methylation in the serum of colorectal cancer patients. Clin Cancer Res. 2002;8:188-91.
35.Noh YH, Im G, Ku JH, Lee YS, Ahn MJ. Detection of tumor cell contamination in peripheral blood by RT-PCR in gastrointestinal cancer patients. J Korean Med Sci. 1999;14:623- 8.
36.Grady WM, Rajput A, Lutterbaugh JD, Markowitz SD. Detection of aberrantly methylated hMLHl promoter DNA in the serum of patients with microsatellite unstable colon cancer. Cancer Res. 2001 ;61:900-2.
37.Nakayama H, Hibi K, Taguchi M, Takase T, Yamazaki T, Kasai Y, et al. Molecular detection of p16 promoter methylation in the serum of colorectal cancer patients. Cancer Lett. 2002;188:115-9.
38.Verma M, Srivastava S. Epigenetics in cancer: implications for early detection and prevention. Lancet Oncol. 2002;3:755-63.
00.Xx M, Chen WD, Papadopoulos N, Goodman SN, Bjerregaard NC, Laurberg S, et al. Sensitive digital quantification of DNA methylation in clinical samples. Nat Biotechnol. 2009;27:858-63.
40.Grutzmann R, Molnar B, Pilarsky C, Habermann JK, Schlag PM, Saeger HD, et al. Sensitive detection of colorectal cancer in peripheral blood by septin 9 DNA methylation assay. PLoS One. 2008;3:e3759. PMCID: 2582436.
41.Lofton-Day C, Model F, Devos T, Tetzner R, Distler J, Schuster M, et al. DNA methylation biomarkers for blood-based colorectal cancer screening. Clin Chem. 2008;54:414- 23.
42.Hofsli E, Sjursen W, Prestvik WS, Johansen J, Rye M, Trano G, et al. Identification of serum microRNA profiles in colon cancer. Br J Cancer. 2013;108:1712-9. PMCID: 3668463.
43.Apweiler R, Hermjakob H, Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta. 1999;1473:4-8.
44.Adamczyk B, Tharmalingam T, Rudd PM. Glycans as cancer biomarkers. Biochim Biophys Acta. 2012;1820:1347-53.
45.Patwa TH, Zhao J, Anderson MA, Simeone DM, Lubman DM. Screening of glycosylation patterns in serum using natural glycoprotein microarrays and multi-lectin fluorescence detection. Anal Chem. 2006;78:6411-21.
46.Yue T, Maupin KA, Fallon B, Li L, Partyka K, Anderson MA, et al. Enhanced discrimination of malignant from benign pancreatic disease by measuring the CA 19-9 antigen on specific protein carriers. PLoS One. 2011;6:e29180. PMCID: 3248411.
47.Zhao J, Patwa TH, Qiu W, Shedden K, Hinderer R, Misek DE, et al. Glycoprotein microarrays with multi-lectin detection: unique lectin binding patterns as a tool for classifying normal, chronic pancreatitis and pancreatic cancer sera. J Proteome Res. 2007;6:1864-74.
48.Zhao J, Qiu W, Simeone DM, Lubman DM. N-linked glycosylation profiling of pancreatic cancer serum using capillary liquid phase separation coupled with mass spectrometric analysis. J Proteome Res. 2007;6:1126-38.
49.Qiu Y, Patwa TH, Xu L, Shedden K, Misek DE, Tuck M, et al. Plasma glycoprotein profiling for colorectal cancer biomarker identification by lectin glycoarray and lectin blot. J Proteome Res. 2008;7:1693-703.
50.Rho JH, Mead JR, Wright WS, Brenner DE, Stave JW, Gildersleeve JC, et al. Discovery of sialyl Lewis A and Lewis X modified protein cancer biomarkers using high density antibody arrays. J Proteomics. 2014;96:291-9. PMCID: 3946870.
51.Bresalier RS, Byrd JC, Tessler D, Lebel J, Koomen J, Hawke D, et al. A circulating ligand for galectin-3 is a haptoglobin-related glycoprotein elevated in individuals with colon cancer. Gastroenterology. 2004;127:741-8.
52.Scanlan MJ, Welt S, Gordon CM, Chen YT, Gure AO, Stockert E, et al. Cancer-related serological recognition of human colon cancer: identification of potential diagnostic and immunotherapeutic targets. Cancer Res. 2002;62:4041-7.
00.Xx H, Goodell V, Disis ML. Targeting serum antibody for cancer diagnosis: a focus on colorectal cancer. Expert Opin Ther Targets. 2007;11:235-44.
54.Nam MJ, Madoz-Gurpide J, Wang H, Lescure P, Schmalbach CE, Zhao R, et al. Molecular profiling of the immune response in colon cancer using protein microarrays: occurrence of autoantibodies to ubiquitin C-terminal hydrolase L3. Proteomics. 2003;3:2108-15.
26
55.Zaenker P, Ziman MR. Serologic autoantibodies as diagnostic cancer biomarkers--a review. Cancer Epidemiol Biomarkers Prev. 2013;22:2161-81.
56.Reipert BM, Tanneberger S, Pannetta A, Bedosti M, Poell M, Zimmermann K, et al. Increase in autoantibodies against Fas (CD95) during carcinogenesis in the human colon: a hope for the immunoprevention of cancer? Cancer Immunol Immunother. 2005;54:1038-42.
57.He Y, Wu Y, Mou Z, Li W, Zou L, Fu T, et al. Proteomics-based identification of HSP60 as a tumor-associated antigen in colorectal cancer. Proteomics Clin Appl. 2007;1:336-42.
58.Chen Y, Lin P, Qiu S, Peng XX, Looi K, Farquhar MG, et al. Autoantibodies to Ca2+ binding protein Calnuc is a potential marker in colon cancer detection. Int J Oncol. 2007;30:1137-44.
59.Liu W, Wang P, Li Z, Xu W, Dai L, Wang K, et al. Evaluation of tumour-associated antigen (TAA) miniarray in immunodiagnosis of colon cancer. Scand J Immunol. 2009;69:57-63.
60.Giovannucci E. Insulin-like growth factor- I and binding protein-3 and risk of cancer. Horm Res. 1999;51:34-41.
61.Giovannucci E, Pollak MN, Platz EA, Willet WC, Stampfer MJ, Majeed N, et al. A prospective study of plasma Insulin-like growth factor and binding protein-3 and risk of colorectal neoplasia in women. Cancer Epidemiol Biomarkers Prev. 2000;9:345-9.
62.Giovannucci E, Pollak M, Platz EA, Willet WC, Stampfer MJ, Majeed N, et al. Insulin- like growth factor I (IGF-1), IGF-binding protein-3 and the risk of colorectal adenoma and cancer in the Nurses’ Health Study. Growth Horm IGF Res. 2000;10 Suppl A:S30-l.
63.Palmquist R, Stattin P, Rinaldi S, Biessy C, Stenling R, Riboli E, et al. Plasma insulin, IGF-binding proteins-I and -2 and risk of colorectal cancer: a prospective study in northern Sweden. Int J Cancer. 2003;107:89-93.
64.Cruz-Correa M, Cui H, Giardiello FM, Powe NR, Hylind L, Robinson A, et al. Loss of imprinting of insulin growth factor II gene: a potential heritable biomarker for colon neoplasia predisposition. Gastroenterology. 2004;126:964-70.
65.Cui H, Onyango P, Brandenburg S, Wu Y, Hsieh CL, Feinberg AP. Loss of imprinting in colorectal cancer linked to hypomethylation of Hl 9 and IGF2. Cancer Res. 2002;62:6442-6.
66.Cui H, Cruz-Correa M, Giardiello FM, Hutcheon DF, Kafonek DR, Brandenburg S, et al. Loss ofIGF2 imprinting: a potential marker of colorectal cancer risk. Science. 2003;299:1753-5.
67.Woodson K, Flood A, Green L, Tangrea JA, Hanson J, Cash B, et al. Loss of insulin-like growth factor-II imprinting and the presence of screen-detected colorectal adenomas in women. J Natl Cancer Inst. 2004;96:407-10.
68.Tsushima H, Kawata S, Tamura S, Ito N, Shirai Y, Kiso S, et al. High levels of transforming growth factor beta 1 in patients with colorectal cancer: association with disease progression. Gastroenterology. 1996;110:375-82.
69.Tsushima H, Ito N, Tamura S, Matsuda Y, Inada M, Yabuuchi I, et al. Circulating Transforming Growth Factor beta-I as a predictor of liver metastasis after resection in colorectal cancer. Clin Cancer Res. 2001;7:1258-62.
70.Narai S, Watanabe M, Hasegawa H, Nishibori H, Endo T, Kubota T, et al. Significance of Transforming gro’-¾1h factor beta 1 as a new tumor marker for colorectal cancer. Int J Cancer. 2002;97:508-11.
71.Broll R, Erdmann H, Duchrow M, Oevermann E, Schwandner 0, Markert U, et al. Vascular endothelial growth factor (VEGF) -- a valuable serum tumour marker in patients with colorectal cancer? Eur J Surg Oncol. 2001;27:37-42.
72.Takeda A, Shimada H, Imaseki H, Okazumi S, Natsume T, Suzuki T, et al. Clinical significance of serum vascular endothelial growth factor in colorectal cancer patients : correlation with clinicopathological factors and tumor markers. Oncol Rep. 2000;7:333-8.
73.Shimoyama S, Yamasaki K, Kawahara M, Kaminishi M. Increased serum angiogenin concentration in colorectal cancer is correlated with cancer progression. Clin Cancer Res. 1999;5:1125-30.
74.Feldman AL, Alexander HR, Jr, Bartlett DL, Kranda KC, Miller MS, Costouros NG, et al. A prospective analysis of plasma endostatin levels in colorectal cancer patients with liver metastases. Ann Surg Oncol. 2001;8:741-5.
75.Simpson RA, Dickinson T, Porter KE, London NJ, Hemingway DM. Raised levels of plasma big endothelin 1 in patients with colorectal cancer. Br J Surg. 2000;87:1409-13.
76.Peeters CF, Thomas CM, Sweep FC, Span PN, Wobbes T, Ruers TM. Elevated serum endothelin-1 levels in patients with colorectal cancer; relevance for prognosis. Int J Biol Markers. 2000;15:288-93.
77.Pellegrini P, Contasta I, Berghella AM, Gargano E, Mammarella C, Adorno D. Simultaneous measurement of soluble carcinoembryonic antigen and the tissue inhibitor of metalloproteinase TIMP 1 serum levels for use as markers of pre-invasive to invasive colorectal cancer. Cancer Immunol Immunother. 2000;49:388-94.
78.Yukawa N, Yoshikawa T, Akaike M, Sugimasa Y, Takemiya S, Yanoma S, et al. Plasma concentration of tissue inhibitor of matrix metalloproteinase 1 in patients with colorectal carcinoma. Br J Surg. 2001;88:1596-601.
79.Barozzi C, Ravaioli M, D’Errico A, Grazi GL, Poggioli G, Cavrini G, et al. Relevance of biologic markers in colorectal carcinoma: a comparative study of a broad panel. Cancer. 2002;94:647-57.
80.Holten-Anderson MN, Christensen IJ, Nielsen HJ, Stephens RW, Jensen V, Nielsen OH, et al. Total levels of tissue inhibitor of metalloproteinases 1 in plasma yield high diagnostic sensitivity and specificity in patients with colon cancer. Clin Cancer Res. 2002;8:156-64.
27
81.Alexiou 0, Karayiannakis AJ, Syrigos KN, Zbar A, Kremmyda A, Bramis I, et al. Serum levels of E-selectin, ICAM-1 and VCAM-1 in colorectal cancer patients: correlations with clinicopathological features, patient survival and tumour surgery. Eur J Cancer. 2001;37:2392-7.
82.Hayes OF, Smerage JB. Circulating tumor cells. Prog Mol Biol Transl Sci. 2010;95:95- 112.
83.Wicha MS, Hayes DF. Circulating tumor cells: not all detected cells are bad and not all bad cells are detected. J Clin Oncol. 2011;29:1508-11.
84.Lim SH, Becker TM, Chua W, Ng WL, de Souza P, Spring KJ. Circulating tumour cells and the epithelial mesenchymal transition in colorectal cancer. J Clin Pathol. 2014;67:848-53.
85.Zhang Z, Nagrath S. Microfluidics and cancer: are we there yet? Biomed Microdevices. 2013;15:595-609. PMClD: 4017600.
86.Stott SL, Lee RJ, Nagrath S, Yu M, Miyamoto OT, Ulkus L, et al. Isolation and characterization of circulating tumor cells from patients with localized and metastatic prostate cancer. Sci Transl Med. 2010;2:25ra3. PMCIO: 3141292.
87.Murlidhar V, Zeinali M, Grabauskiene S, Ghannad-Rezaie M, Wicha MS, Simeone DM, et al. A radial flow microfluidic device for ultra-high-throughput affinity-based isolation of circulating tumor cells. Small. 2014;10:4895-904.
88.Cooper DN. Galectinomics: finding themes in complexity. Biochim Biophys Acta. 2002;1572:209-31.
89.Dudas SP, Yunker CK, Sternberg LR, Byrd JC, Bresalier RS. Expression of human intestinal mucin is modulated by the beta-galactoside binding protein galectin-3 in colon cancer. Gastroenterology. 2002;123:817-26.
90.Mazurek N, Conklin J, Byrd JC, Raz A, Bresalier RS. Phosphorylation of the beta- galactoside-binding protein galectin-3 modulates binding to its ligands. J Biol Chem. 2000;275:36311-5.
91.Schoeppner H, Raz A, Ho S, Bresalier R. Expression of an endogenous galactose-binding lectin correlates with neoplastic progression in the colon. Cancer. 1995;75:2818-26.
92.Sanjuan X, Fernandez PL, Castells A, Castronovo V, van den Brule F, Liu FT, et al. Differential expression of galectin 3 and galectin 1 in colorectal cancer progression. Gastroenterology. 1997;113:1906-15.
93.Lotz MM, Andrews CW, Jr., Korzelius CA, Lee EC, Steele GD, Jr., Clarke A, et al. Decreased expression of Mac-2 (carbohydrate binding protein 35) and loss of its nuclear localization are associated with the neoplastic progression of colon carcinoma. Proc Natl Acad Sci US A. 1993;90:3466-70.
94.Levi Z, Rozen P, Hazazi R, Vilkin A, Waked A, Maoz E, et al. A quantitative immunochemical fecal occult blood test for colorectal neoplasia. Ann Intern Med. 2007;146:244- 55.
95.Shastri YM, Stein J. Quantitative immunochemical fecal occult blood test for diagnosing colorectal neoplasia. Ann Intern Med. 2007;147:522-3; author reply 3.
96.Rabeneck L, Rumble RB, Thompson F, Mills M, Oleschuk C, Whibley A, et al. Fecal immunochemical tests compared with guaiac fecal occult blood tests for population-based colorectal cancer screening. Can J Gastroenterol. 2012;26:131-47. PMCID: 3299236.
97.Whitlock EP, Lin JS, Liles E, Beil TL, Fu R. Screening for colorectal cancer: a targeted, updated systematic review for the U.S. Preventive Services Task Force. Ann Intern Med. 2008;149:638-58.
98.Auge JM, Pellise M, Escudero JM, Hernandez C, Andreu M, Grau J, et al. Risk stratification for advanced colorectal neoplasia according to fecal hemoglobin concentration in a colorectal cancer screening program. Gastroenterology. 2014;147:628-36 el.
99.Ahlquist DA, Shuber AP. Stool screening for colorectal cancer: evolution from occult blood to molecular markers. Clin Chim Acta. 2002;3 l 5:157-68.
100.Osborn NK, Ahlquist DA. Stool screening for colorectal cancer: molecular approaches. Gastroenterology. 2005;128:192-206.
101.Ahlquist DA, Sargent DJ, Loprinzi CL, Levin TR, Rex DK, Ahnen DJ, et al. Stool DNA and occult blood testing for screen detection of colorectal neoplasia. Ann Intern Med. 2008;149:441-50, W81.
102.lmperiale TF, Ransohoff DF, Itzkowitz SH, Turnbull BA, Ross ME. Fecal DNA versus fecal occult blood for colorectal-cancer screening in an average-risk population. N Engl J Med. 2004;351:2704-14.
103.Imperiale TF, Ransohoff DF, Itzkowitz SH, Levin TR, Lavin P, Lidgard GP, et al. Multitarget Stool DNA Testing for Colorectal-Cancer Screening. N Engl J Med. 2014.
104.Force USPST. Final Research Plan: Colorectal Cancer Screening 2014. Available from: xxxx://xxx.xxxxxxxxxxxxxxxxxxxxxxxxxxxxx .org/Page/Document/ResearchPlanFina l/colorectal cancer-screening2.
105.Chen WD, Han ZJ, Skoletsky J, Olson J, Sah J, MyeroffL, et al. Detection in fecal DNA of colon cancer-specific methylation of the nonexpressed vimentin gene. J Natl Cancer Inst. 2005;97:1124-32.
106.Zou H, Harrington JJ, Shire AM, Rego RL, Wang L, Campbell ME, et al. Highly methylated genes in colorectal neoplasia: implications for screening. Cancer Epidemiol Biomarkers Prev. 2007;16:2686-96.
107.Itzkowitz S, Brand R, Jandorf L, Durkee K, Millholland J, Rabeneck L, et al. A simplified, noninvasive stool DNA test for colorectal cancer detection. Am J Gastroenterol. 2008;103:2862-70.
108.Itzkowitz SH, Jandorf L, Brand R, Rabeneck L, Schroy PC, 3rd, Sontag S, et al. Improved fecal DNA test for colorectal cancer screening. Clin Gastroenterol Hepatol. 2007;5:l 1l-7.
109.Zackular JP, Rogers MA, Ruffin MTt, Schloss PD. The human gut microbiome as a screening tool for colorectal cancer. Cancer Prev Res (Phila). 2014;7:1112-21. PMCID: 4221363.
28
110.Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207-15. PMCID: 3772512.
111.Montrose DC, Zhou XK, Kopelovich L, Yantiss RK, Karoly ED, Subbaramaiah K, et al. Metabolic profiling, a noninvasive approach for the detection of experimental colorectal neoplasia. Cancer Prev Res (Phila). 2012;5:1358-67. PMCID: 3518611.
112.Ahmed FE, Ahmed NC, Vos P\V, Bonnerup C, Atkins JN, Casey M, et al. Diagnostic microRNA markers to screen for sporadic human colon cancer in stool: I. Proof of principle. Cancer Genomics Proteomics. 2013;10:93-113.
113.Link A, Balaguer F, Shen Y, Nagasaka T, Lozano JJ, Boland CR, et al. Fecal MicroR.”T\l”As as novel biomarkers for colon cancer screening. Cancer Epidemiol Biomarkers Prev. 2010;19:1766-74. PMCID: 2901410.
114.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr., Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546-58. PMCID: 3749880.
115.Lieberman DA, Williams JL, Holub JL, Morris CD, Logan JR, Eisen GM, et al. Race, ethnicity, and sex affect risk for polyps >9 mm in average-risk individuals. Gastroenterology. 2014;147:351-8; quiz e14-5. PMCID: 4121117.
116.Lieberman DA, Holub JL, Morris CD, Logan J, Williams JL, Camey P. Low rate of large polyps (>9 mm) within 10 years after an adequate baseline colonoscopy with no polyps. Gastroenterology. 2014;147:343-50.
117.Dominitz JA, Robertson DJ. Tailoring colonoscopic screening to individual risk. Gastroenterology. 2014;147:264-6.
118.Moinova H, Leidner RS, Ravi L, Lutterbaugh J, Barnholtz-Sloan JS, Chen Y, et al. Aberrant vimentin methylation is characteristic of upper gastrointestinal pathologies. Cancer Epidemiol Biomarkers Prev. 2012;21:594-600. PMCID: 3454489.
119.Fedirko V, Bostick RM, Flanders WD, Long Q, Shaukat A, Rutherford RE, et al. Effects of vitamin D and calcium supplementation on markers of apoptosis in normal colon mucosa: a randomized, double-blind, placebo-controlled clinical trial. Cancer Prev Res (Phila Pa). 2009;2:213-23.
120.Weinstein SJ, Albanes D, Selhub J, Graubard B, Lim U, Taylor PR, et al. One-carbon metabolism biomarkers and risk of colon and rectal cancers. Cancer Epidemiol Biomarkers Prev. 2008;17:3233-40. PMCID: 2656360.
121.Giovannucci E, Stampfer M, Golditz G, Hunter D, Fuchs C, Rosner B, et al. Multivitamin use, folate, and colon cancer in women in the nurses’ health study. Ann Intern Med. 1998;129:517-24.
122.Le Marchand L, White KK, Nomura AM, Wilkens LR, Selhub JS, Tiirikainen M, et al. Plasma levels of B vitamins and colorectal cancer risk: the multiethnic cohort study. Cancer Epidemiol Biomarkers Prev. 2009;18:2195-201.
123.Bruce WR, Giacca A, Medline A. Possible mechanisms relating diet and risk of colon cancer. Cancer Epidemiol Biomarkers Prev. 2000;9:1271-9.
124.English DR, Macinnis RJ, Hodge AM, Hopper JL, Haydon AM, Giles GG. Red meat, chicken, and fish consumption and risk of colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2004;13:1509-14.
125.Terry P, Giovannucci E, Michels KB, Bergkvist L, Hansen H, Holmberg L, et al. Fruit, vegetables, dietary fiber, and risk of colorectal cancer. J Natl Cancer Inst. 2001;93:525-33.
126.Trock B, Lanza E, Greenwald P. Dietary fiber, vegetables, and colon cancer: critical review and meta-analyses of the epidemiologic evidence. J Natl Cancer Inst. 1990;82:650-6 1.
127.Vogel V, McPherson R. Dietary epidemiology of colon cancer. Hematol Oncol Clin North Am. 1989;3:35-63.
128.Ribas-Barba L, Serra-Majem L, Roman-Vinas B, Ngo J, Garcia-Alvarez A. Effects of dietary assessment methods on assessing risk of nutrient intake adequacy at the population level: from theory to practice. Br J Nutr. 2009;101 Suppl 2:S64-72.
129.Roman-Vinas B, Serra-Majem L, Ribas-Barba L, Ngo J, Garcia-Alvarez A, Wijnhoven TM, et al. Overview of methods used to evaluate the adequacy of nutrient intakes for individuals and populations. Br J Nutr. 2009;101 Suppl 2:S6-l 1.
130.Lipworth L, Bender TJ, Rossi M, Bosetti C, Negri E, Talamini R, et al. Dietary vitamin D intake and cancers of the colon and rectum: a case-control study in Italy. Nutr Cancer. 2009;61:70-5.
131.Jacobs ET, Thompson PA, Martinez ME. Diet, gender, and colorectal neoplasia. J Clin Gastroenterol. 2007;41:731-46.
132.Forte A, De Sanctis R, Leonetti G, Manfredelli S, Urbano V, Bezzi M. Dietary chemoprevention of colorectal cancer. Ann Ital Chir. 2008;79:261-7.
000.xx Vogel S, Bongaerts BW, Wouters KA, Kester AD, Schouten LJ, de Goeij AF, et al. Associations of dietary methyl donor intake with MLHl promoter hypermethylation and related molecular phenotypes in sporadic colorectal cancer. Carcinogenesis. 2008;29:1765-73.
134.Powers HJ, Hill MH, Welfare M, Spiers A, Bal W, Russell J, et al. Responses of biomarkers of folate and riboflavin status to folate and riboflavin supplementation in healthy and colorectal polyp patients (the FAB2 Study). Cancer Epidemiol Biomarkers Prev. 2007;16:2128- 35.
135.Slattery ML, Potter JD, Duncan DM, Berry TD. Dietary fats and colon cancer: assessment of risk associated with specific fatty acids. Int J Cancer. 1997;73:670-7.
136.Suhar AF, Thompson FE, Kipnis V, Midthune D, Hurwitz P, McNutt S, et al. Comparative validation of the Block, Willett, and National Cancer Institute food frequency questionnaires: the Eating at America’s Table Study. Am J Epidemiol. 2001;154:1089-99.
29
000.Xx FB, Rimm E, Smith-Warner SA, Feskanich D, Stampfer MJ, Ascherio A, et al. Reproducibility and validity of dietary patterns assessed with a food-frequency questionnaire. Am J Clin Nutr. 1999;69:243-9.
138.Bloch DA. Comparing two diagnostic tests against the same “gold standard” in the same sample. Biometrics. 1997;53:73-85.
139.Lieberman DA, Rex DK, Winawer SJ, Giardiello FM, Johnson DA, Levin TR. Guidelines for colonoscopy surveillance after screening and polypectomy: a consensus update by the US Multi-Society Task Force on Colorectal Cancer. Gastroenterology. 2012;143:844-57.
140.Ahlquist DA, Zou H, Domanico M, Mahoney DW, Yab TC, Taylor WR, et al. Next generation stool DNA test accurately detects colorectal cancer and large adenomas. Gastroenterology. 2012;142:248-56; quiz e25-6.
141.Bantis LE, Feng Z. Comparison of two correlated ROC curves at a given specificity or sensitivity level. Stat Med. 2016;35:4352-67.
142.Lloyd C, Yong Z. Kernel estimators of the ROC curve are better than emipircal Statistics and Probability Letters. 1999;44.
30